| id | total_length_mm |
|---|---|
| 001-Po-16/05/10 | 213 |
| 002-Po-29/05/10 | 124 |
| 003-Pd-29/05/10 | 166 |
Week 2: Data visualization
EVR 628- Intro to Environmental Data Science
Rosenstiel School of Marine, Atmospheric, and Earth Science and Institute for Data Science and Computing
Announcements
- C + J symposium:
- Dec 11-12, but register soon
- Possibility of contributed talks
- IDSC Data Science Fellowship:
- Deadline for 2025-2026 is Tuesday, September 30, 2025, by 5:00 PM
Learning Objectives
By the end of this week, you should be able to:
- Identify types of data
- Use correct jargon when referring to data
- Know that some types of visualizations are better suited for different types of data
- Build a figure using
ggplot2
Today’s class
- Common ground
- Data jargon
- Types of data
- Common visualization goals and charts
- Principles of visualization
Common ground
Some Jargon
- Variable: A measurable quality, quantity or property
- Value: The state of the variable
- Observation: A data point to which variables and values are assigned
- Tabular data: The way we store and represent information. Allows us to relate a variable (column) to an observation (row)
Example: Lionfish Biometry
data_lionfish, from theEVR628toolspackage- Contains biometric measurements for 109 lionfish (Pterois volitans) captured off Puerto Aventuras (Mexico)
Tip
Use ?data_lionfish to look at the documentation.
Example: Lionfish Biometry
- Which of these is a variable?
- A value of that variable?
- How many observations?
Types of Data
The type of data you have can influence how you visualize them
- Numerical: (AKA “quantitative”) a measurement expressed in numbers, instead of words
- Discrete numerical: Counts, integer numbers
- Continuous numerical: Think decimal numbers
- Categorical: (AKA “qualitative”) a measurement expressed in words, instead of numbers
- Ordinal: A special type of categorical data, where the order of the categories has a meaning
Examples: Lionfish Biometry
| id | site | total_length_mm | size_class |
|---|---|---|---|
| 001-Po-16/05/10 | Paraiso | 213 | large |
| 002-Po-29/05/10 | Paraiso | 124 | medium |
| 003-Pd-29/05/10 | Pared | 166 | medium |
| 004-Cs-12/06/10 | Canones | 203 | large |
| 005-Cs-12/06/10 | Canones | 212 | large |
| 006-Pl-21/06/10 | Paamul | 210 | large |
What type of data is shown in variable id?
How about total_length_mm?
And size_class?
Example: Hurricane Tracks
| name | iso_time | lat | lon | sshs |
|---|---|---|---|---|
| MILTON | 2024-10-05 09:00:00 | 21.7 | -95.5 | -3 |
| MILTON | 2024-10-05 12:00:00 | 22.0 | -95.5 | -1 |
| MILTON | 2024-10-05 15:00:00 | 22.3 | -95.5 | -1 |
| MILTON | 2024-10-05 18:00:00 | 22.5 | -95.5 | 0 |
| MILTON | 2024-10-05 21:00:00 | 22.6 | -95.5 | 0 |
What type of data is sshs?
Trick question!
Use ?data_milton to look at the documentation.
What would -2.5 in sshs mean?
Common visualization goals and charts
Many Types of Visualizations
There are many, many, MANY types of visualizations:
- scatter plot, column chart (stacked columns, dodge columns), histogram, 2d-histogram, violin plots, boxplots, pie charts (eww), heatmaps, line graphs, area charts, stacked area, density, voronoi plots, alluvial diagrams…
But what really matters:
- You want your type of data to match your type of visualization
- Some data types may be “incompatible” with some visualizations
- You should have creative freedom
- We have one goal: To communicate something to the viewer (our ourselves)
Visualizing for Yourself
Right Visuals for Your Goal
Remember our goal:
To communicate something to the viewer (our ourselves)
That something is usually one of 4:
Distribution: “Most of my fish are quite small” (example)
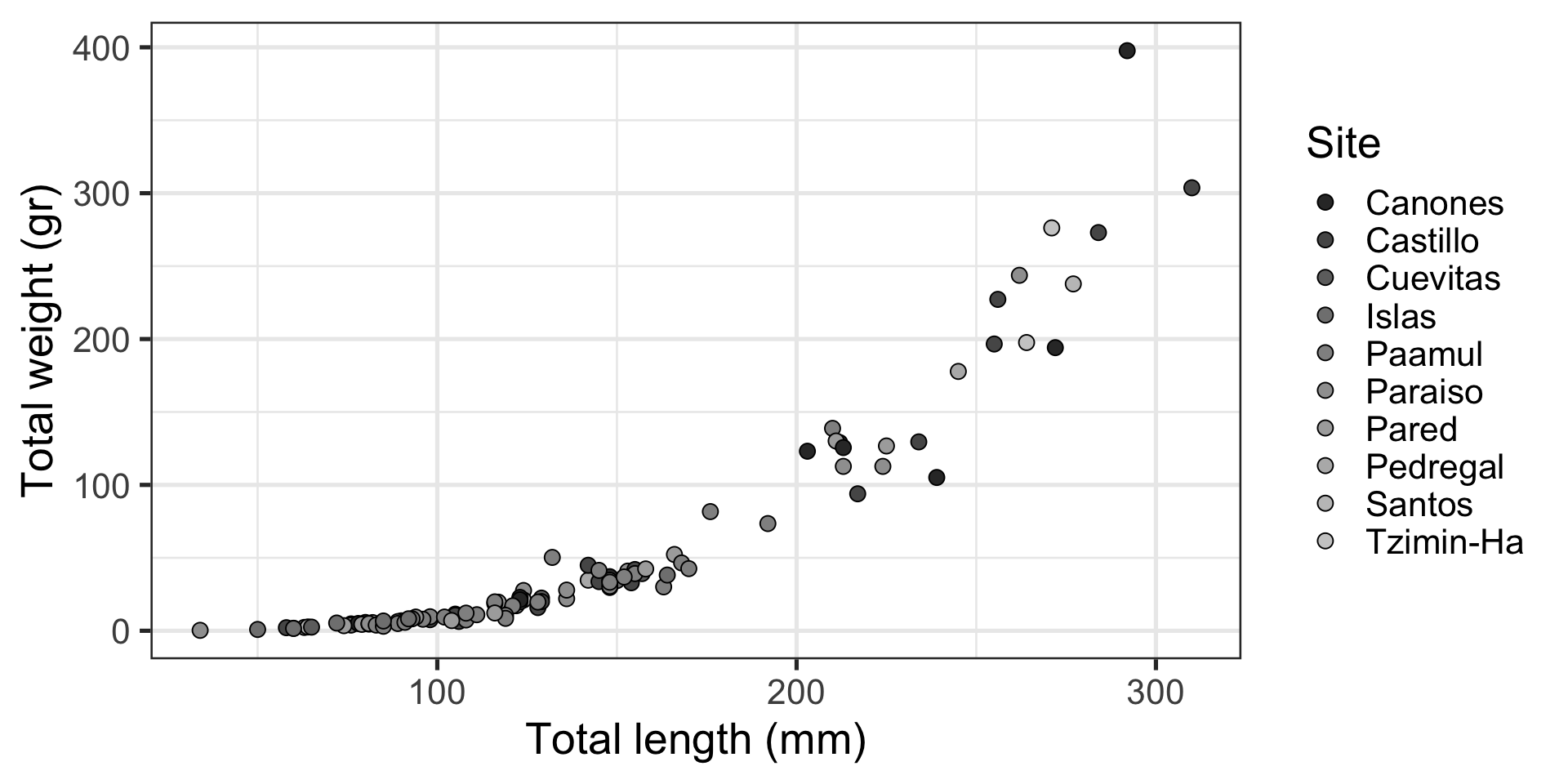
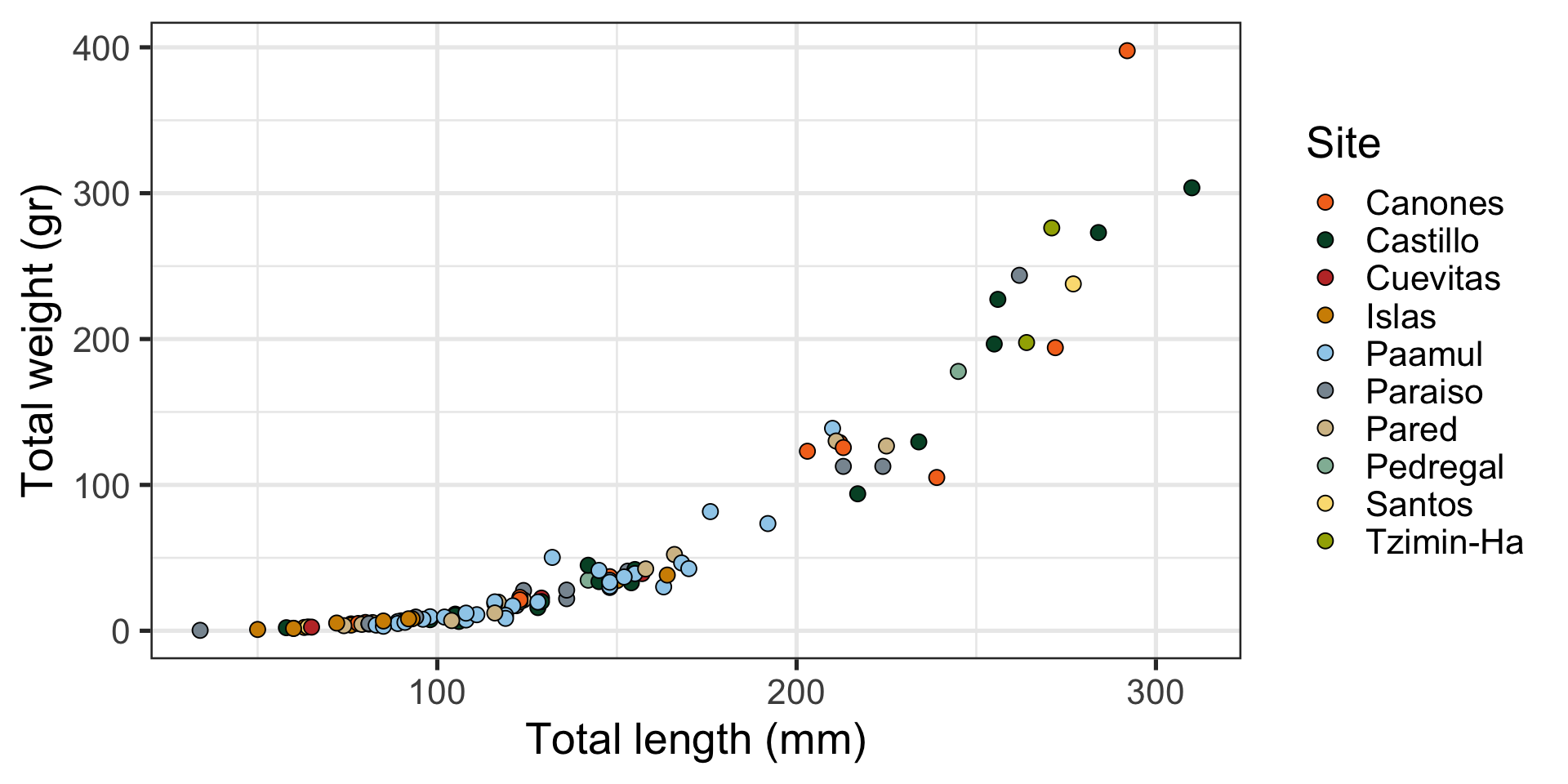
Relationship (or correlation): “Look, slightly larger fish are waaaay heavier!” (example)
Evolution: “The wind speed of a hurricane was highest the night of Oct 7th”(example)
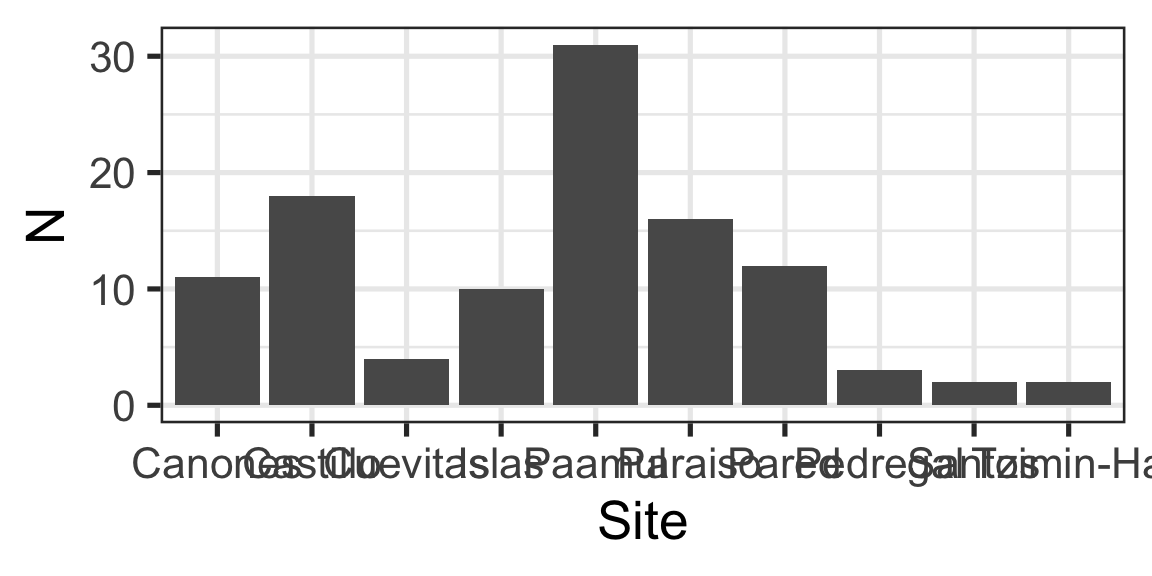
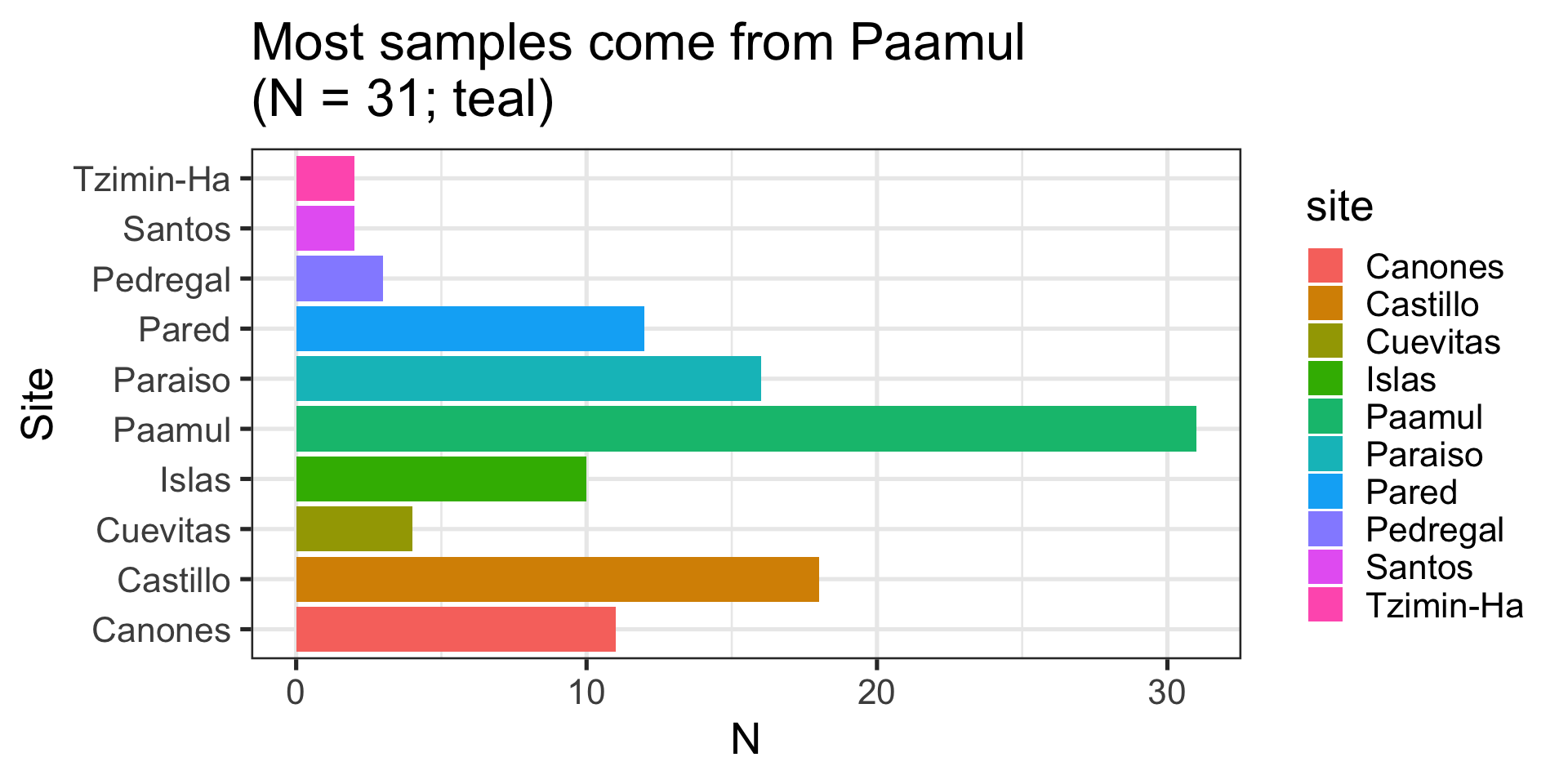
Ranking / Part of a whole: “Largest fish comes from Castillo”(example)
Any visual can usually achieve more than one of this at a time
Most visuals can only achieve one of these effectively at a time
Examples
Code
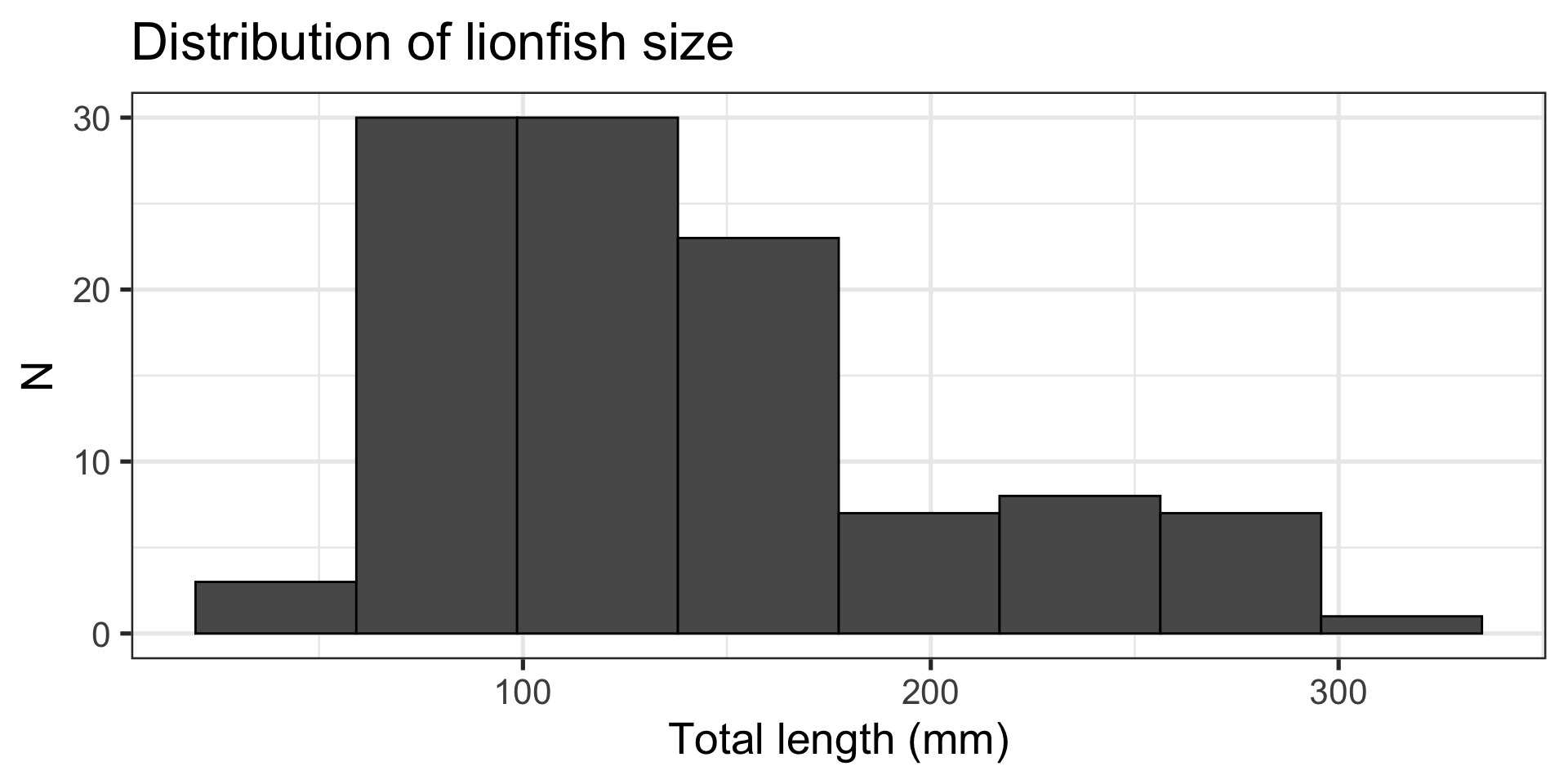
Message: “Most lionfish are around 100 mm in length”
Histogram:
- Shows two numeric variables
- Binned continuous
- Counts within each bin
- Shows the distribution of the data
- Built with
ggplot2::geom_histogram()
Examples
Code
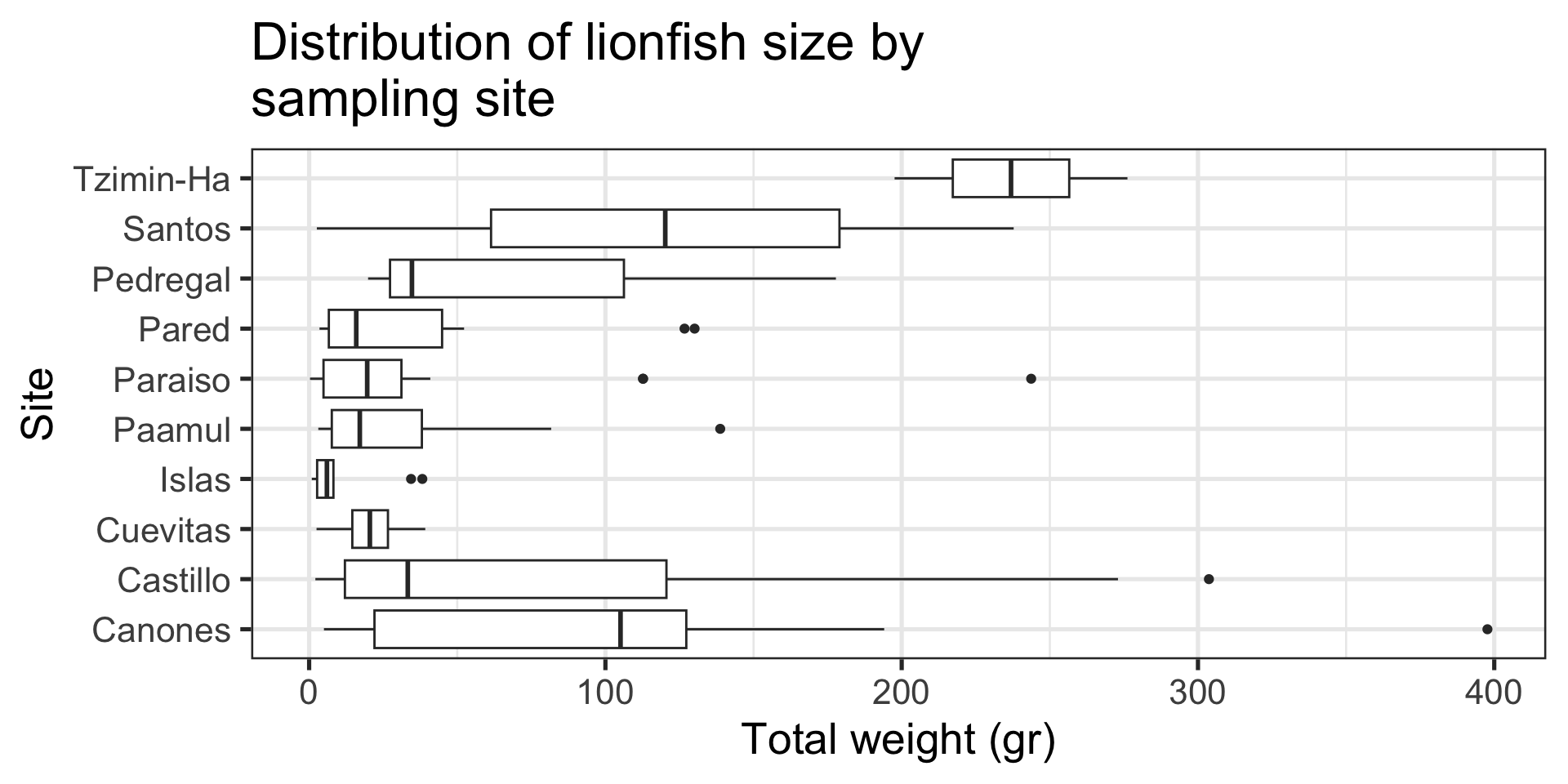
Message: “Tzimin-Ha has the heaviest fish”
Boxplot:
- Distribution of a continuous variable by groups
- Line inside box shows median
- Outlines of box show 25th and 75th percentiles
- Whiskers extend from hinge to the largest value within \(1.5 \times IQR\) from the hinge
- Points are “outlying” observations
Tip
Use this one with care. It packs a lot of information and not everyone knows (remembers) how to read it.
Examples
Code
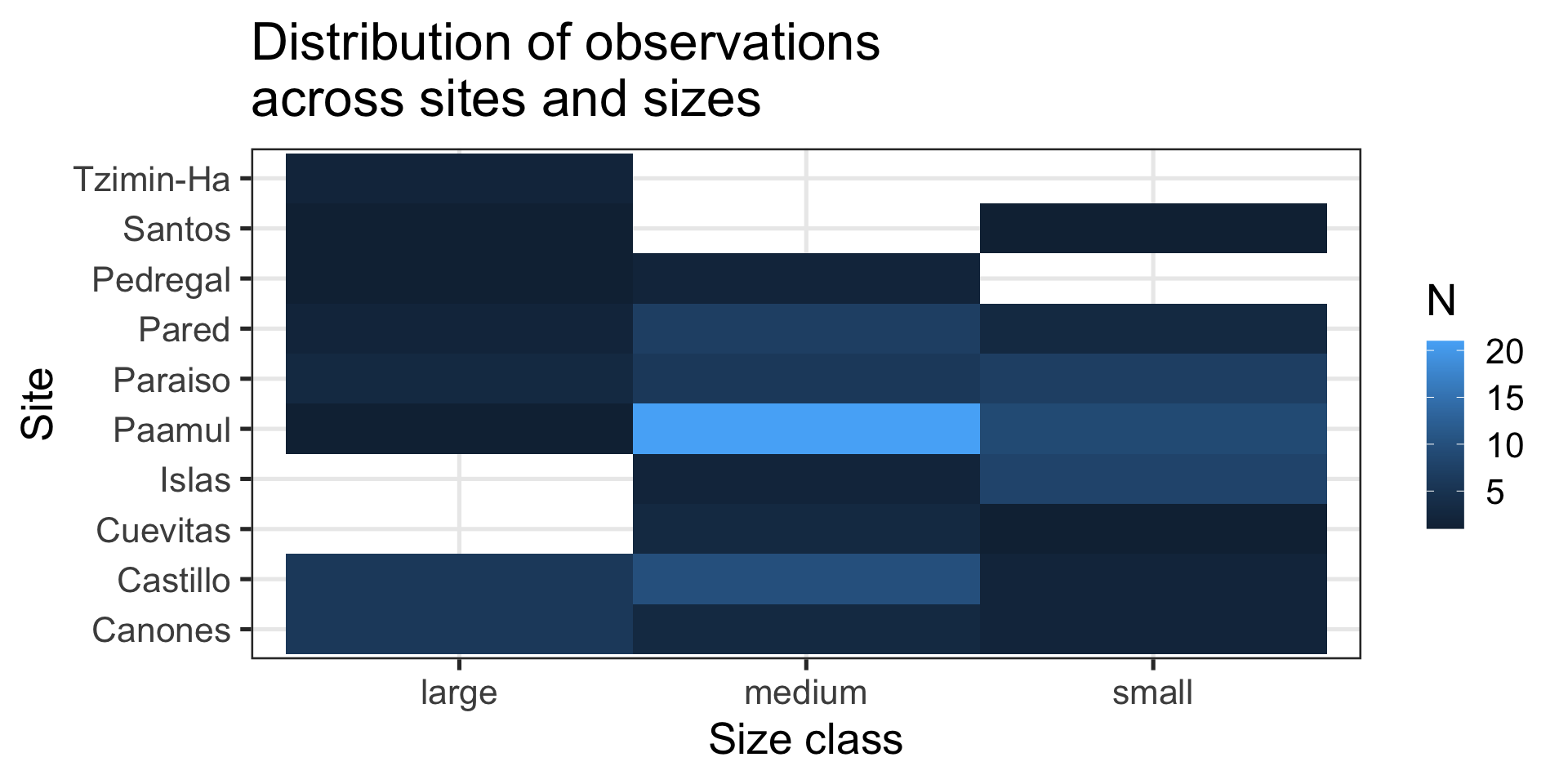
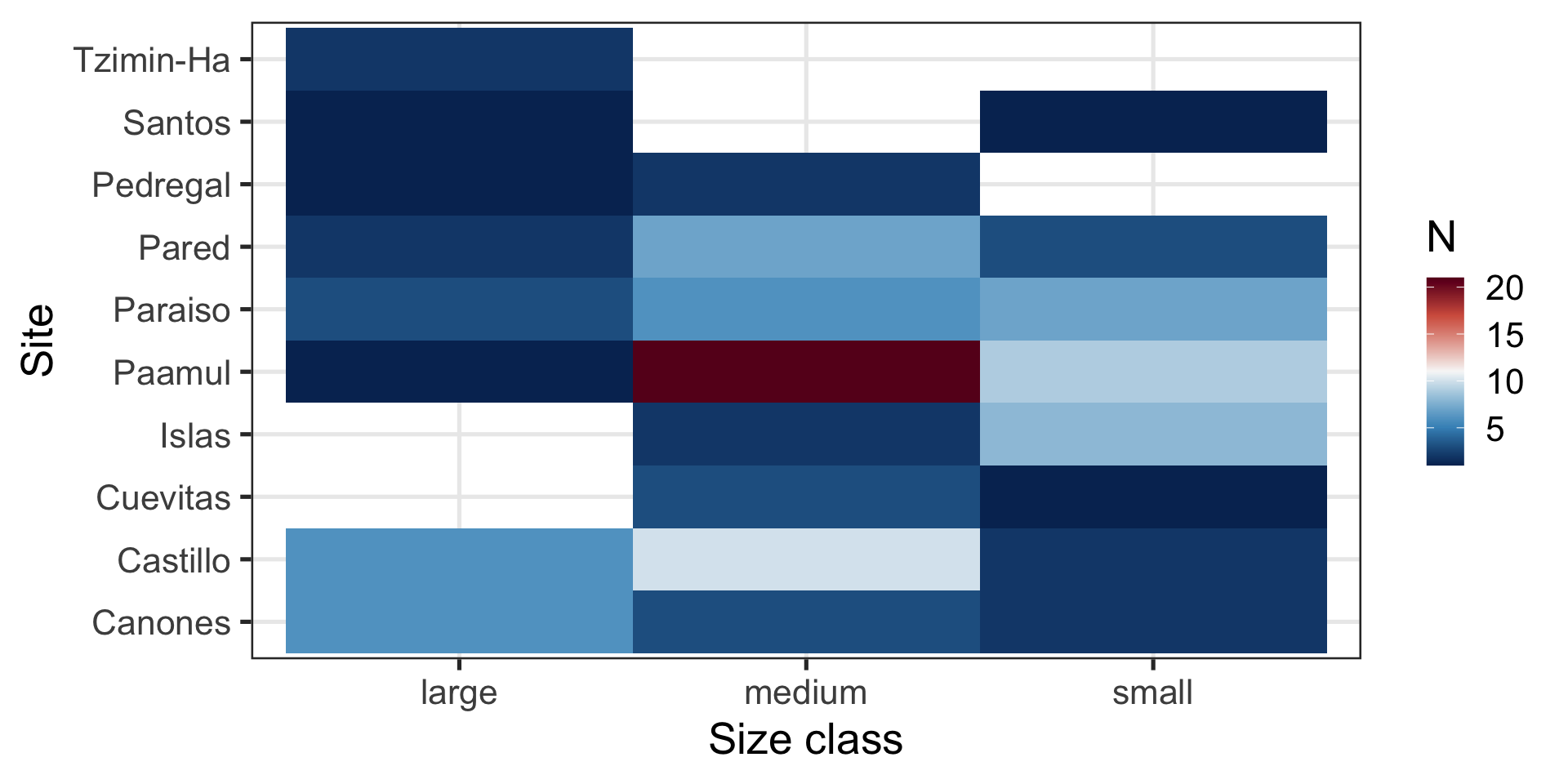
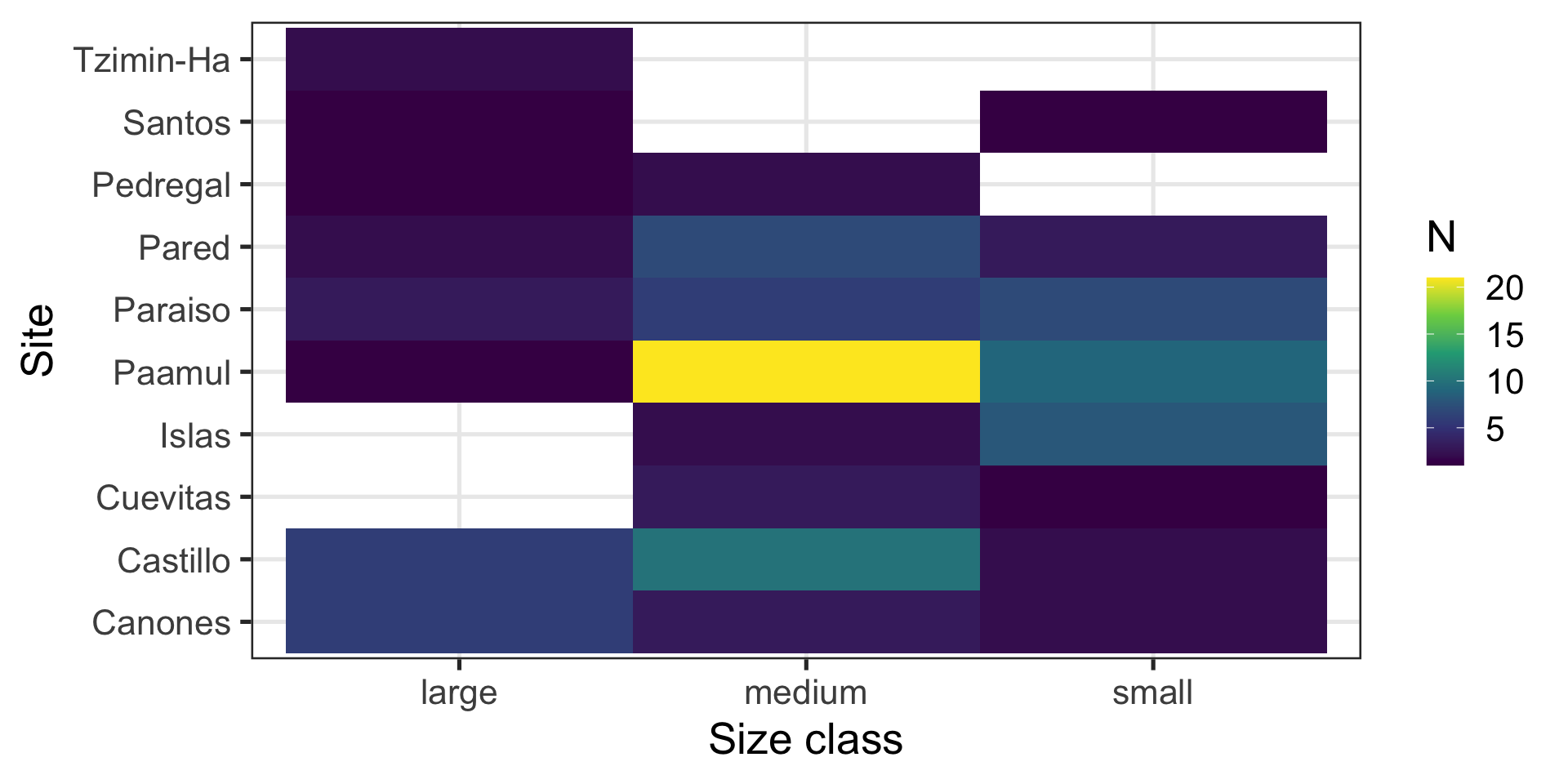
Message: “Most fish are medium sized and come from Paamul”
Heatmap:
- Shows counts across two variables at the same time
- Uses
ggplot2::geom_bin_2d()
Is my x-axis bothering you?
R does not know that there is a logical order (small -> medium -> large).
Examples
Code
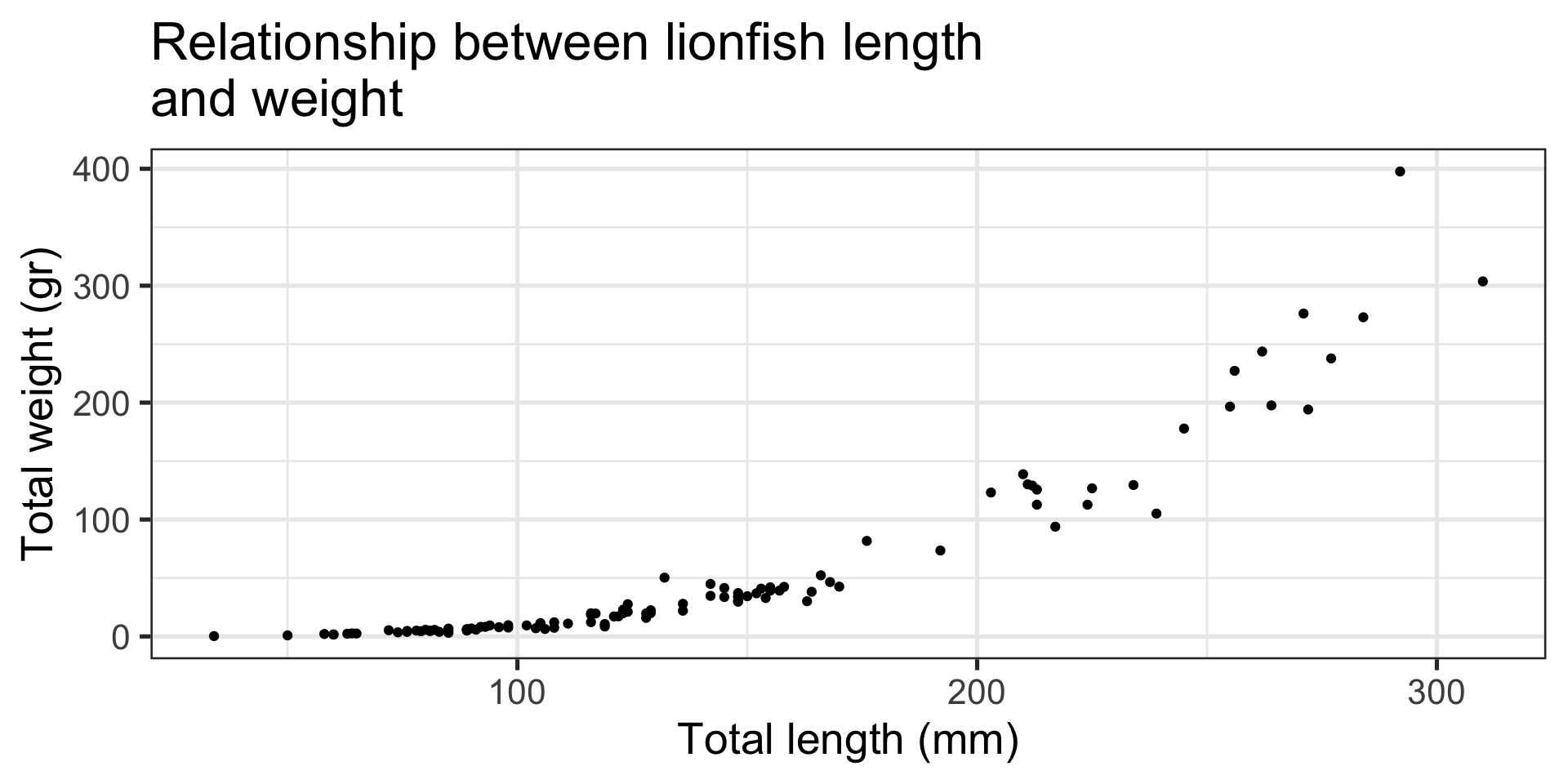
Message: “Look, slightly larger fish are waaaay heavier!”
Scatterplot:
- Relationship between two continuous numeric variables
- Represented with points
- Uses
ggplot2::geom_point()
Examples
Code
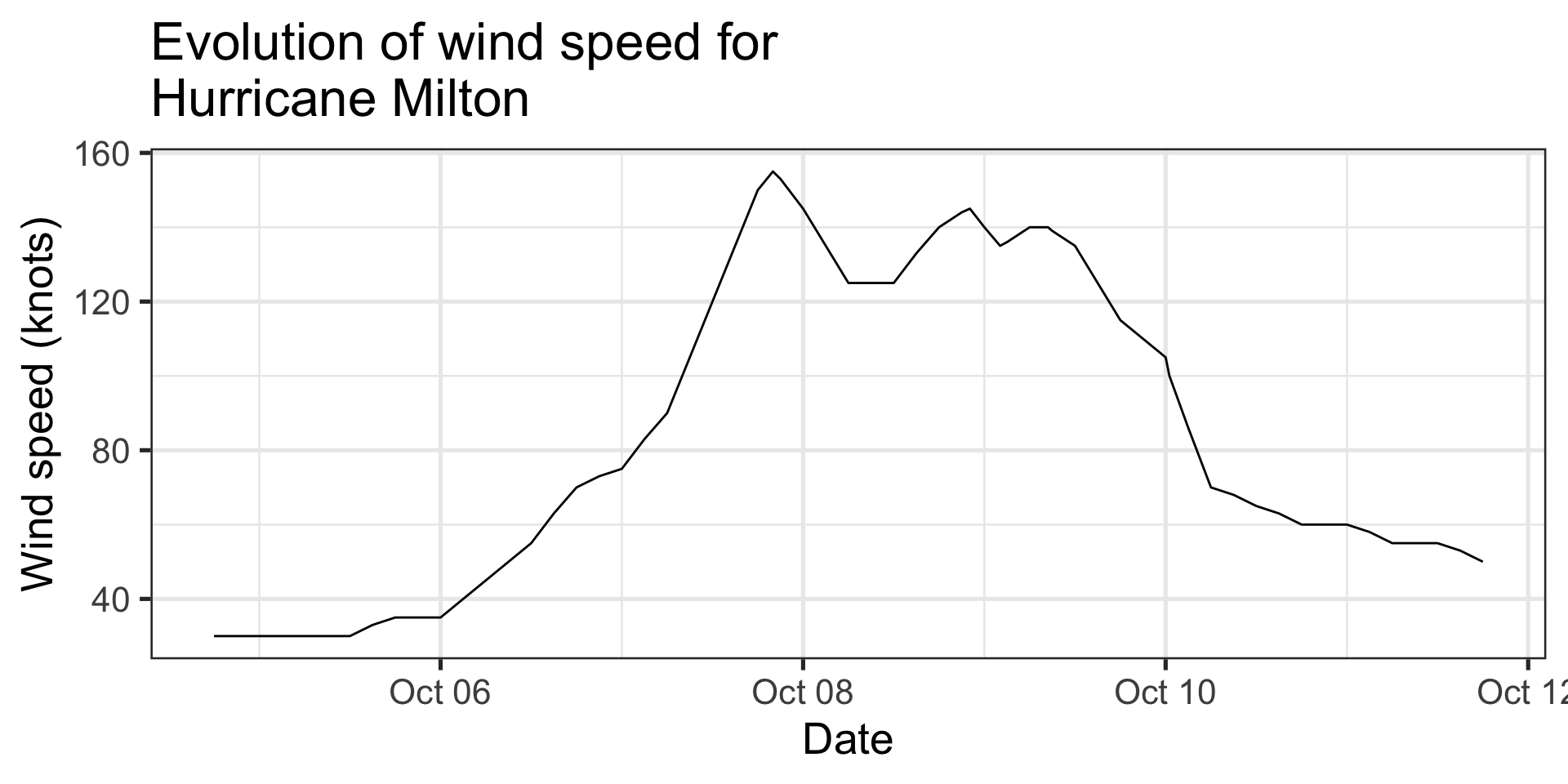
Message: “The wind speed of a hurricane was highest the night of Oct 7th”
Line chart:
- Evolution of one variable along another
- Conveys a sense of continuity even if our observations are not
- Uses
ggplot::geom_line()
Examples
Code
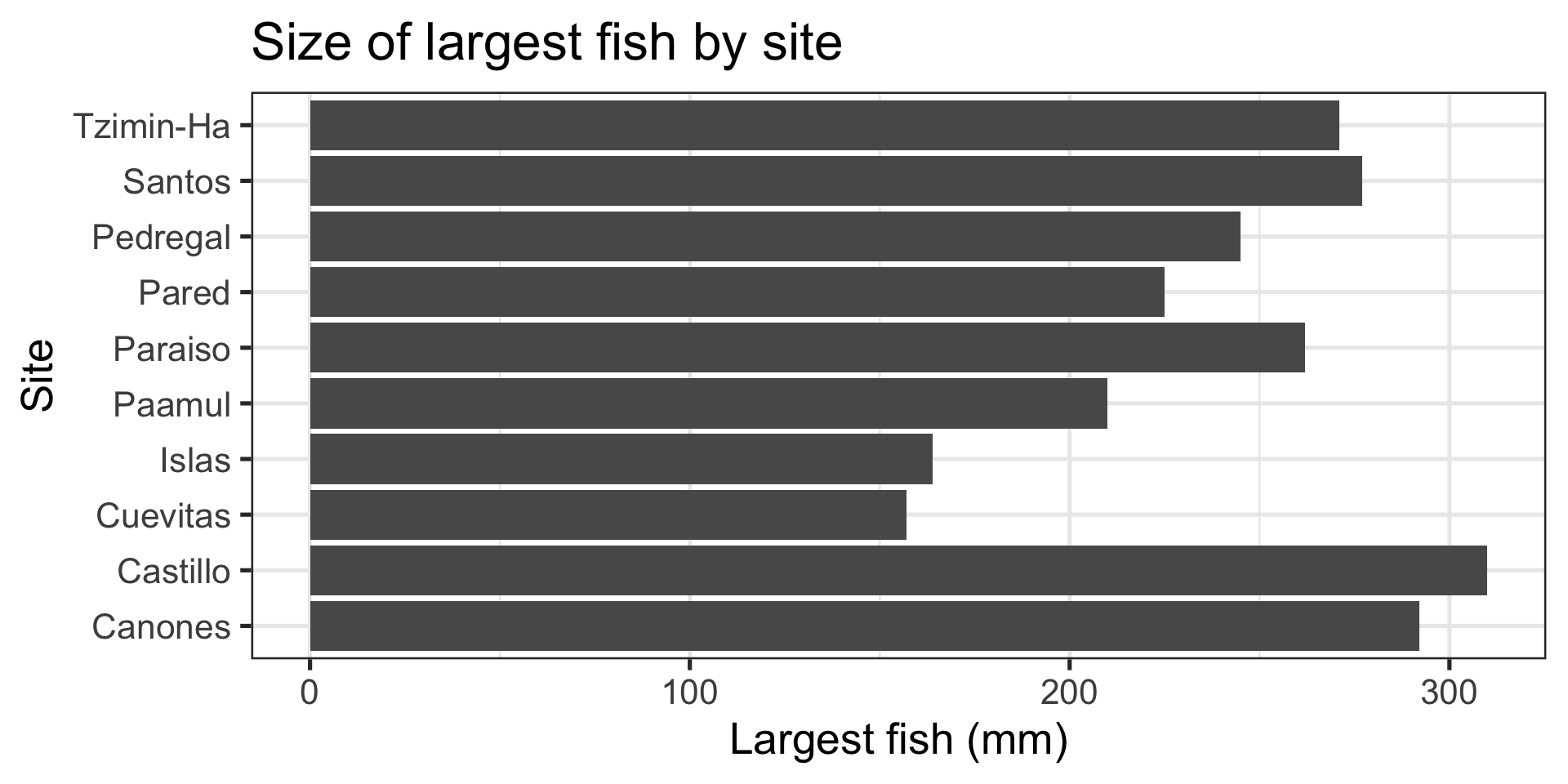
Message: “Largest fish comes from Castillo”
Column:
- Relationship between categorical and numeric variable
- Represented with “columns” or “bars”
- This case uses
ggplot2::geom_col(), but there is alsoggplot2::geom_bar()
Now I’m confused…
- Good!
- That confusion might be critical thinking
- You can always try visualizing the same data in more than one way
Same Data, Multiple Plots
Which is best at showing the relationship between size and weight?
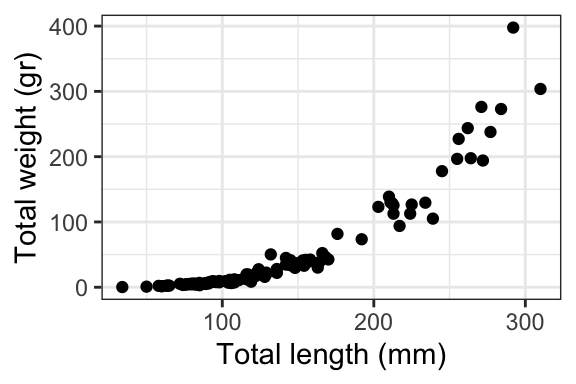
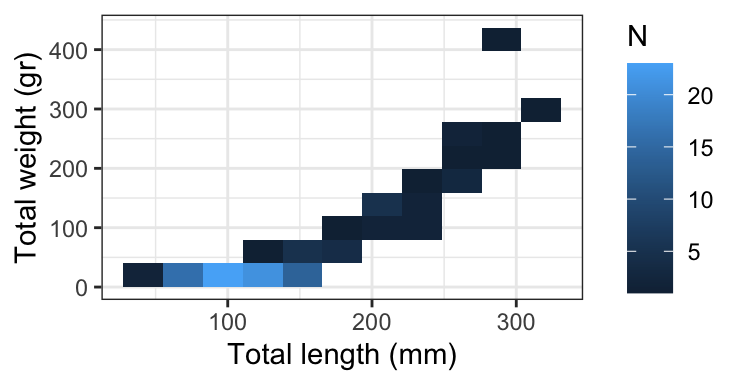
Which is best at showing the size and weight of most fish?
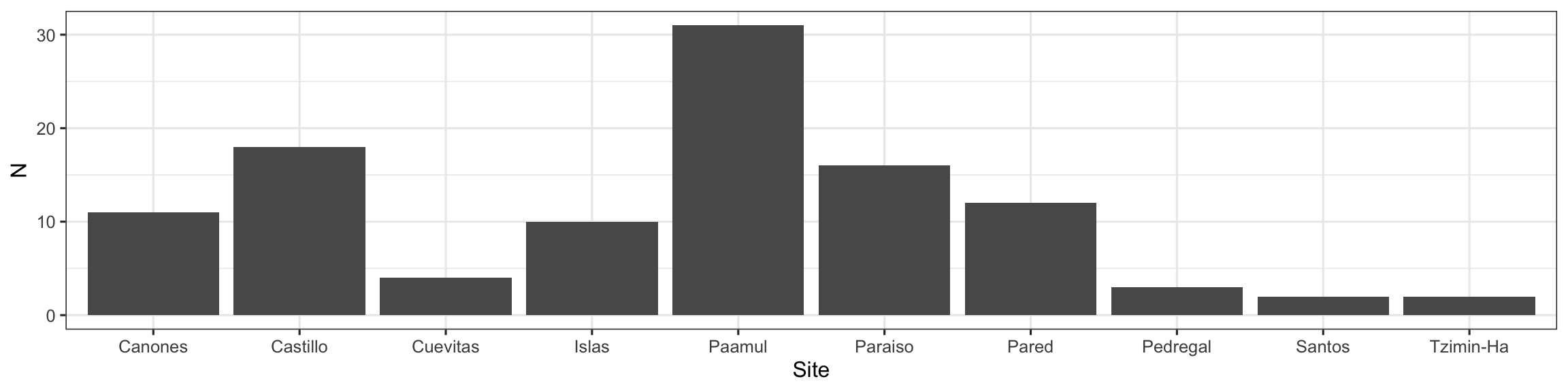
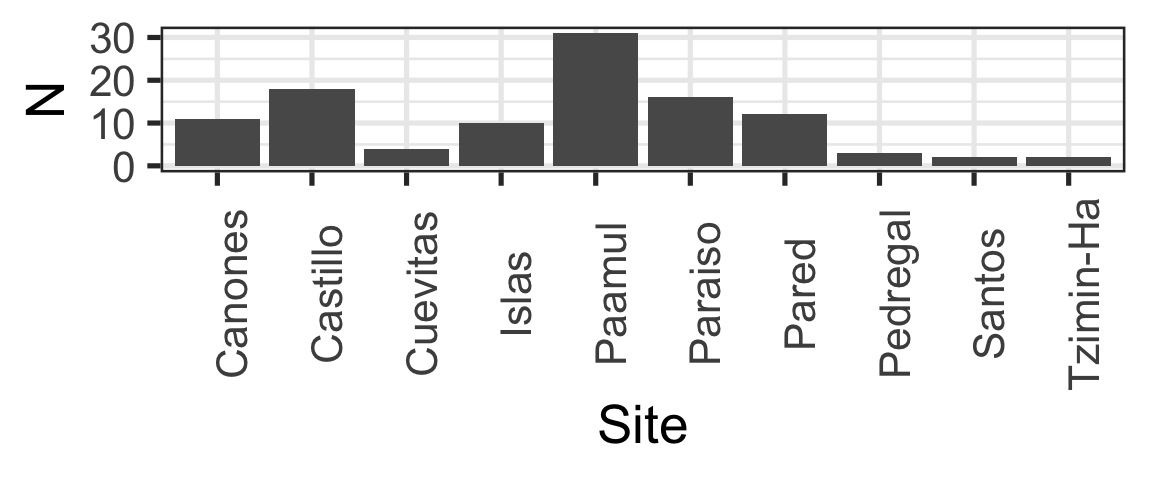
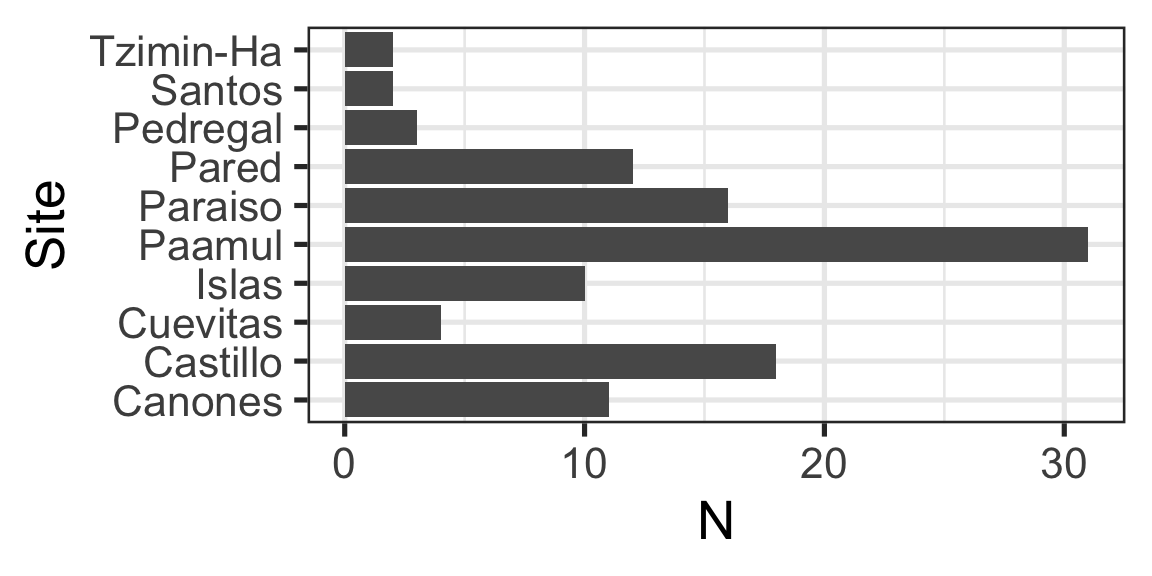
Same Data, Multiple Plots
Which is best at showing me the number of samples by site and size?
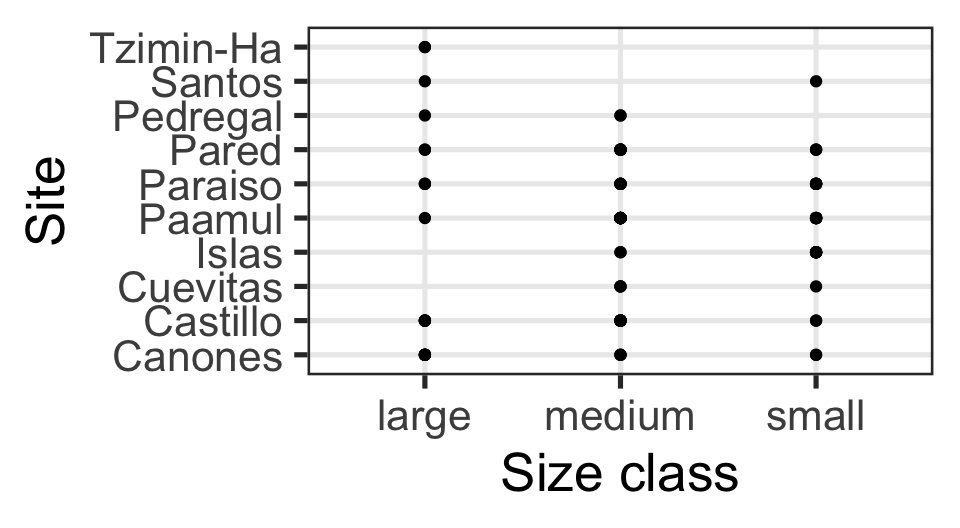
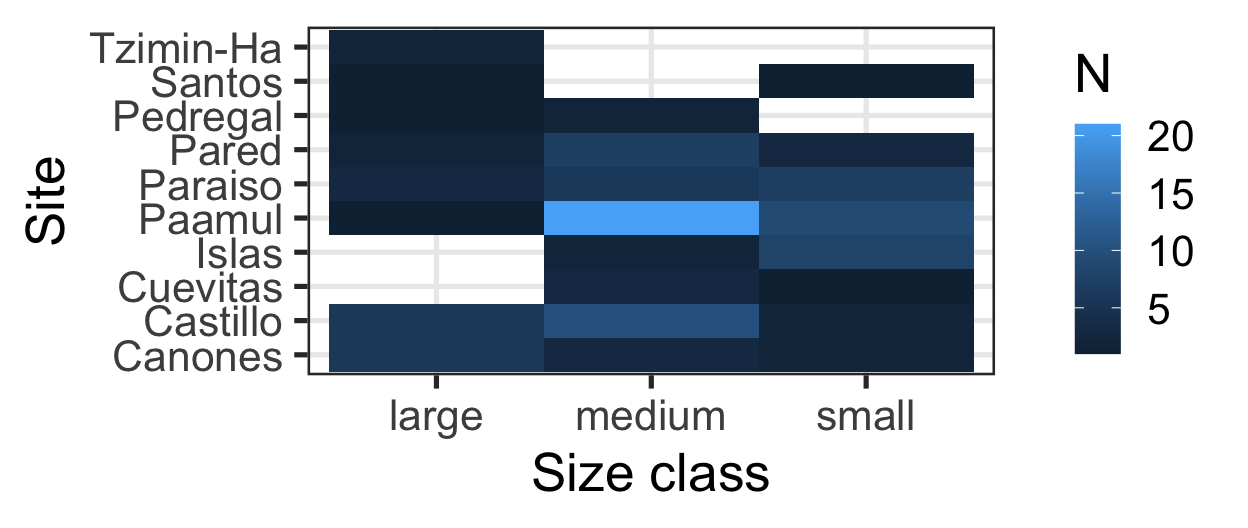


Choosing the “Right” Visual
The right visual is the one that gets the message across
Ask yourself some questions:
- How many variables do I want to represent?
- What type of data do I have for each variable?
- What pattern or message do I want to convey?
There are resources to help you brainstorm:
- r-graph-gallery
- data-to-viz
- What others in your field do
Principles of visualization
You know what type of graph you want, lets make it effective
Jambor (2025)’s Guide to Visualization
- Simplify: Improving annotations, labels, and layouts helps you retain the audience’s attention
- Text: Appropriate font size, avoid non-horizontal text. Axis labels with units!!
- Color scheme:
- Prioritize accessibility and consistency
- Don’t abuse color; Use only when you must draw attention
- If you absolutely need color, consider type of data:
- categorical: Use a discrete color palette (e.g.
EVR628tools::palette_UM()) - ordinal and numeric: single hue with varying saturation or multihue designed for sequential (e.g.
viridiscolors) - If ordinal but diverging data: two contrasting hues and neutral center is better than a single hue or multihues
- categorical: Use a discrete color palette (e.g.
Simplify
Which one is better? Why?
Code
p <- ggplot(data = data_lionfish,
mapping = aes(x = total_length_mm,
y = total_weight_gr)) +
geom_point()
p +
theme_gray() +
scale_x_continuous(breaks = seq(0, 350, by = 15),
limits = c(0, 350)) +
scale_y_continuous(breaks = seq(0, 400, by = 20),
limits = c(0, 400)) +
theme(axis.line = element_line(color = "black"),
panel.grid = element_line(color = "black")) +
labs(title = "Total length (mm) vs total weight (gr) for 109 lionfish sampled from Mexico",
subtitle = "Note that the largest fish is not the heaviest fish")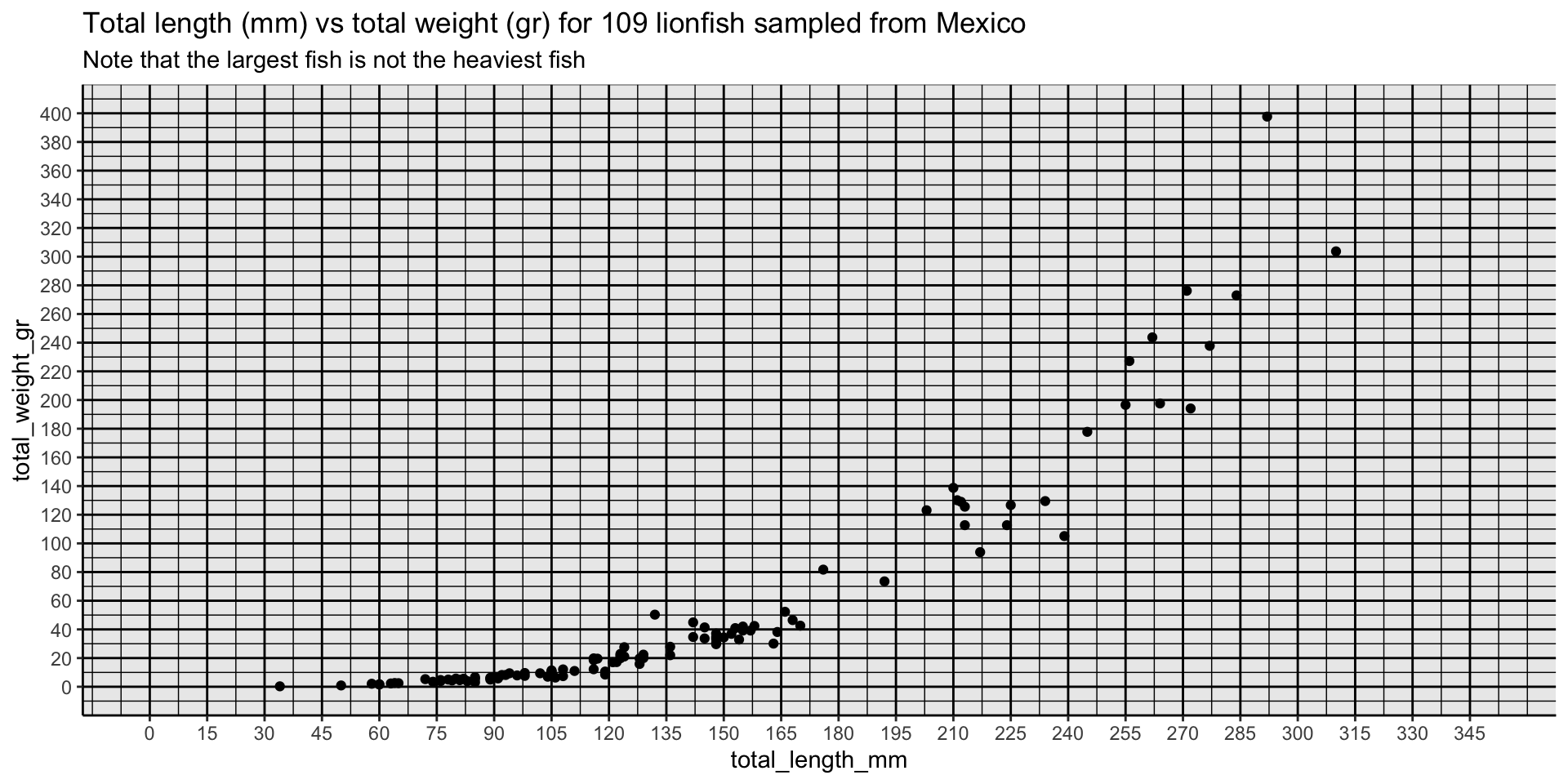
Code
longest <- data_lionfish |> slice_max(total_length_mm)
heaviest <- data_lionfish |> slice_max(total_weight_gr)
p +
geom_text_repel(data = longest,
label = "Longest",
nudge_x = -5,
nudge_y = -150,
size = 5) +
geom_text_repel(data = heaviest,
label = "Heaviest",
nudge_x = -50,
nudge_y = -10,
size = 5) +
theme_minimal(base_size = 14) +
theme(axis.text = element_text(color = "black", size = 10),
axis.title = element_text(color = "black", size = 12)) +
labs(x = "Total length (mm)",
y = "Total weight (gr)") +
labs(title = "There's always a bigger fish",
subtitle = "The largest fish is not the heaviest fish")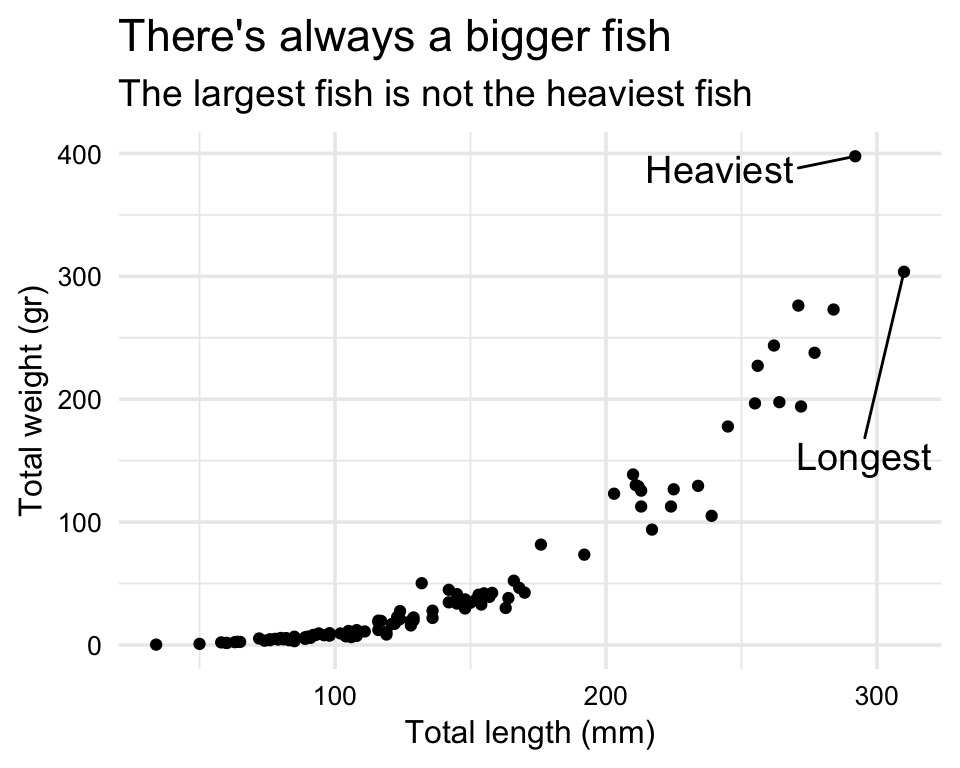
Text
Color Scheme
Code
ggplot(data = data_lionfish,
mapping = aes(x = site, fill = site == "Paamul")) +
geom_bar() +
scale_fill_manual(values = c("FALSE" = "gray",
"TRUE" = "darkred")) +
coord_flip() +
labs(x = "Site", y = "N",
title = "N = 31 come from <span style='color:darkred;'>Paamul</span>") +
theme(plot.title = element_markdown(),
axis.title.y = element_markdown(),
legend.position = "None") +
scale_y_continuous(expand = c(0, 0),
limits = c(0, 32))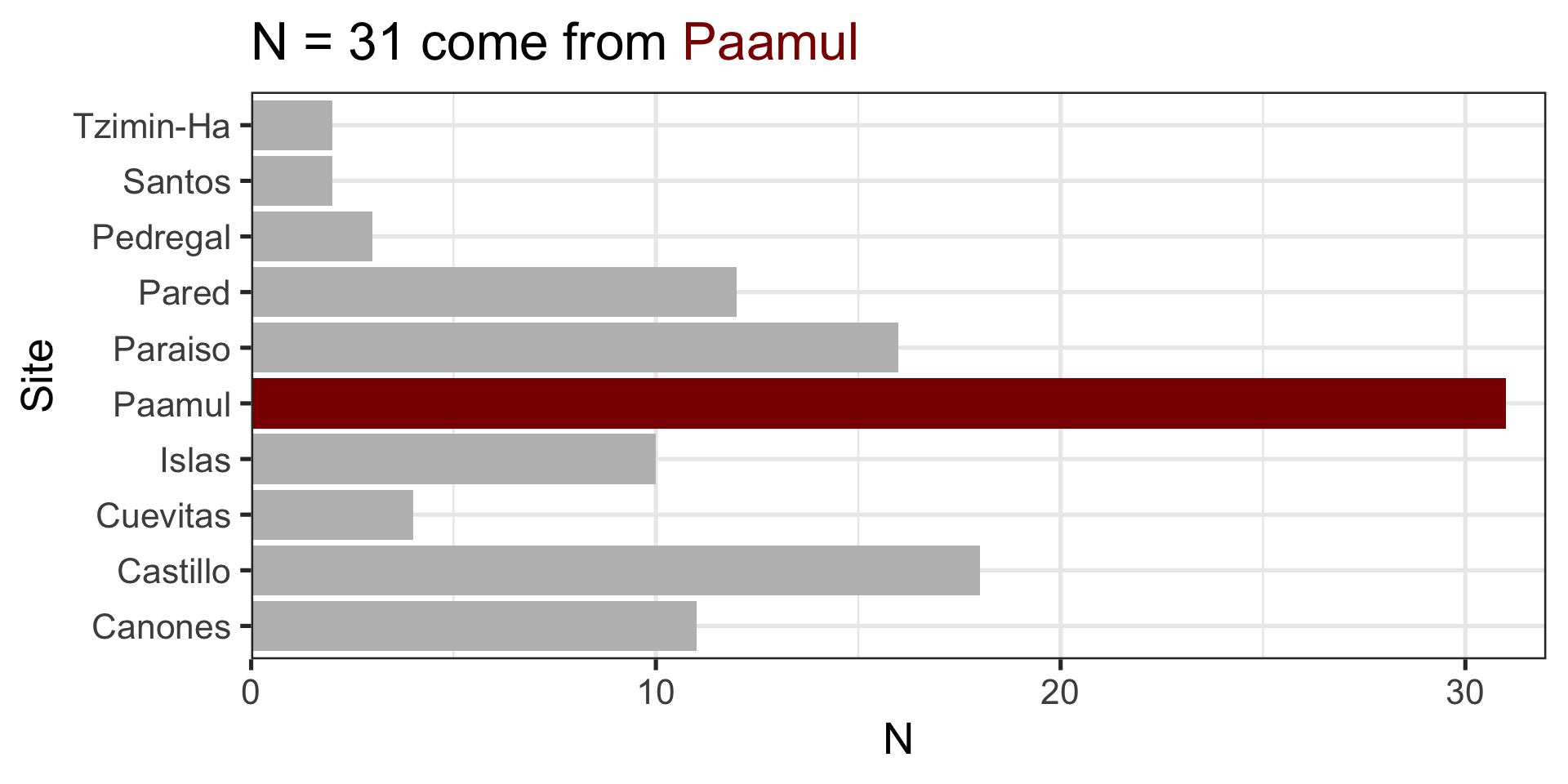
Do you really need to use color?
Use better colors
Avoid redundant use of your limited aesthetics (x, y, size, color, shape)
Categorical data
Use a discrete color palette for categorical data
It is difficult to track more than (6) 10 colors
These colors come from EVR628tools::palette_UM(), using UM’s visual style guide
Ordinal or numeric
Diverging
When was it warmer / colder / average?
Single hue palette
Other Ways of Representing Information
- We’ve tried position (horizontal and vertical) and color
- There is also:
- size
- shape
- aspect (line width, line type)
- Our brains struggle to compare sizes
- We can only track ~6 shapes at a time
- Avoid using these for your most important message
Code
ggplot(data = data_lionfish,
mapping = aes(x = total_length_mm, y = total_weight_gr, color = depth_m, size = fct_relevel(size_class, c("small", "medium", "large")), shape = site)) +
geom_point() +
labs(x = "Total length (mm)",
y = "Total weight (gr)",
color = "Site",
shape = "Site",
size = "Size class") +
scale_shape_manual(values = c(1:10))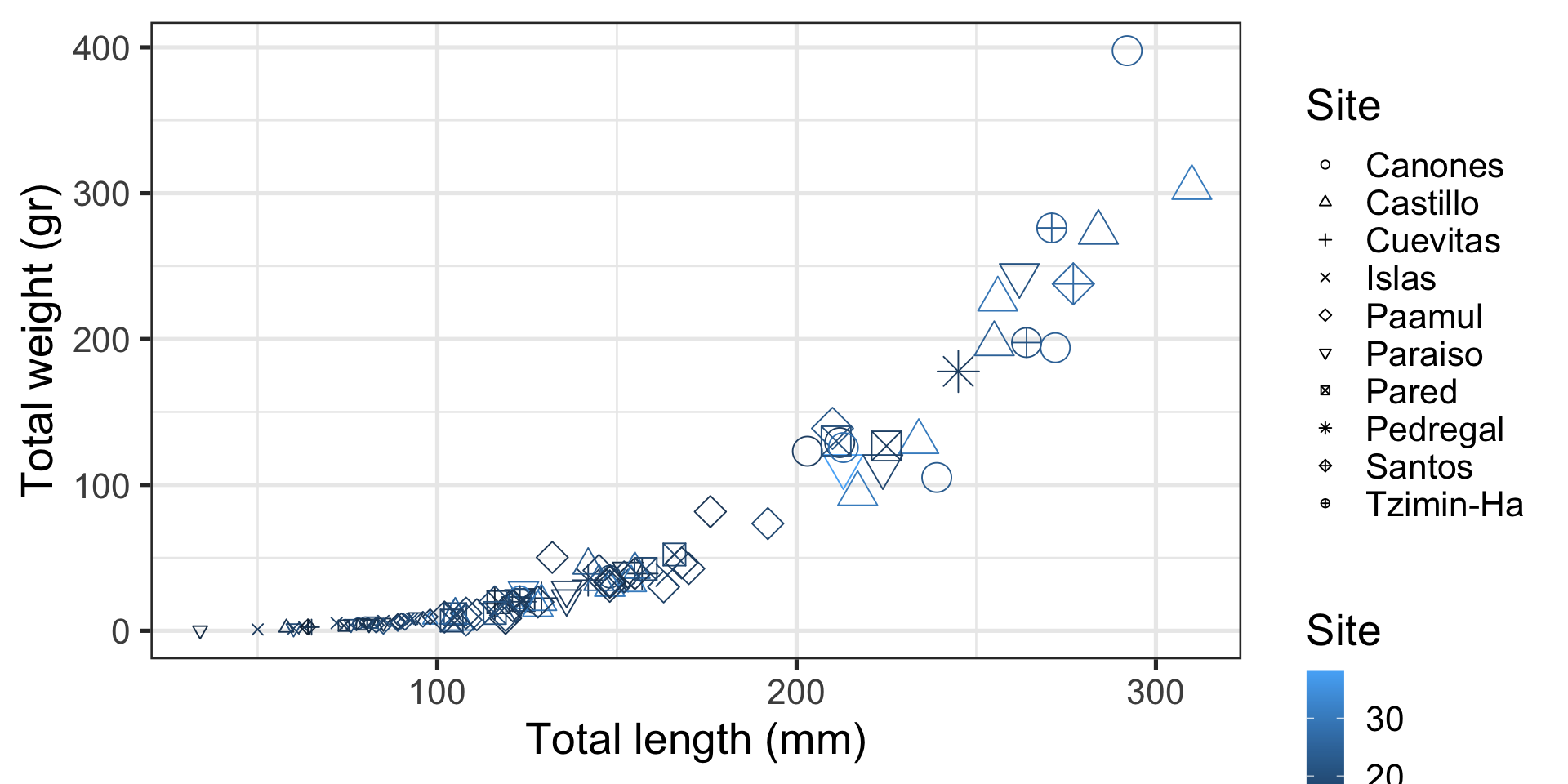
Resources on Style
- A checklist for designing and improving the visualization of scientific data by Jambor (2025)
- r-graph-gallery
- data-to-viz
Use these as guidelines, not as absolute truths
Learning Objectives - Revisited
By the end of this week, you should be able to:
- Identify types of data
- Use correct jargon when referring to data
- Know that some types of visualizations are better suited for different types of data
- Build a figure using
ggplot2
Before Thursday: Read Chapter 1 of R4DS
Intro to ggplot2
The Grammar of Graphics
Grammar:
- Set of rules that define components of a language
Grammar of graphics:
- Proposed by Leland Wilkinson in 2005
- Framework that enables description of components of any graph
- It is NOT a framework for effective visualization
ggplot2:
- Developed by Hadley Wickham
- Leverages the layered grammar of graphics proposed in Wickham (2010)
- Focuses on a layered approach to describe and construct graphs
- Most of the time you will work with
aesthetic mappings andgeometric objects
Requirements
You will need to load two packages
Goal
Recall the data_lionfish data:
Rows: 109
Columns: 9
$ id <chr> "001-Po-16/05/10", "002-Po-29/05/10", "003-Pd-29/05/10…
$ site <chr> "Paraiso", "Paraiso", "Pared", "Canones", "Canones", "…
$ lat <dbl> 20.48361, 20.48361, 20.50167, 20.47694, 20.47694, 20.5…
$ lon <dbl> -87.22611, -87.22611, -87.21167, -87.23278, -87.23278,…
$ total_length_mm <dbl> 213, 124, 166, 203, 212, 210, 132, 122, 224, 117, 211,…
$ total_weight_gr <dbl> 112.70, 27.60, 52.30, 123.10, 129.00, 138.75, 50.29, 1…
$ size_class <chr> "large", "medium", "medium", "large", "large", "large"…
$ depth_m <dbl> 38.1, 27.9, 18.5, 15.5, 15.0, 22.7, 13.4, 18.5, 18.2, …
$ temperature_C <dbl> 28, 28, 28, 28, 28, 29, 29, 29, 29, 29, 28, 28, 28, 28…Goal
Steps to Build a Plot with ggplot2
- Specify your data
- Specify your x (and y) axis
aesthetic mappings - Specify your
geometric representation - Modify
geoms as needed (optional) - Modify your labels as needed
1. Specify the Data
2. Specify the aesthetics
2. Specify the aesthetics
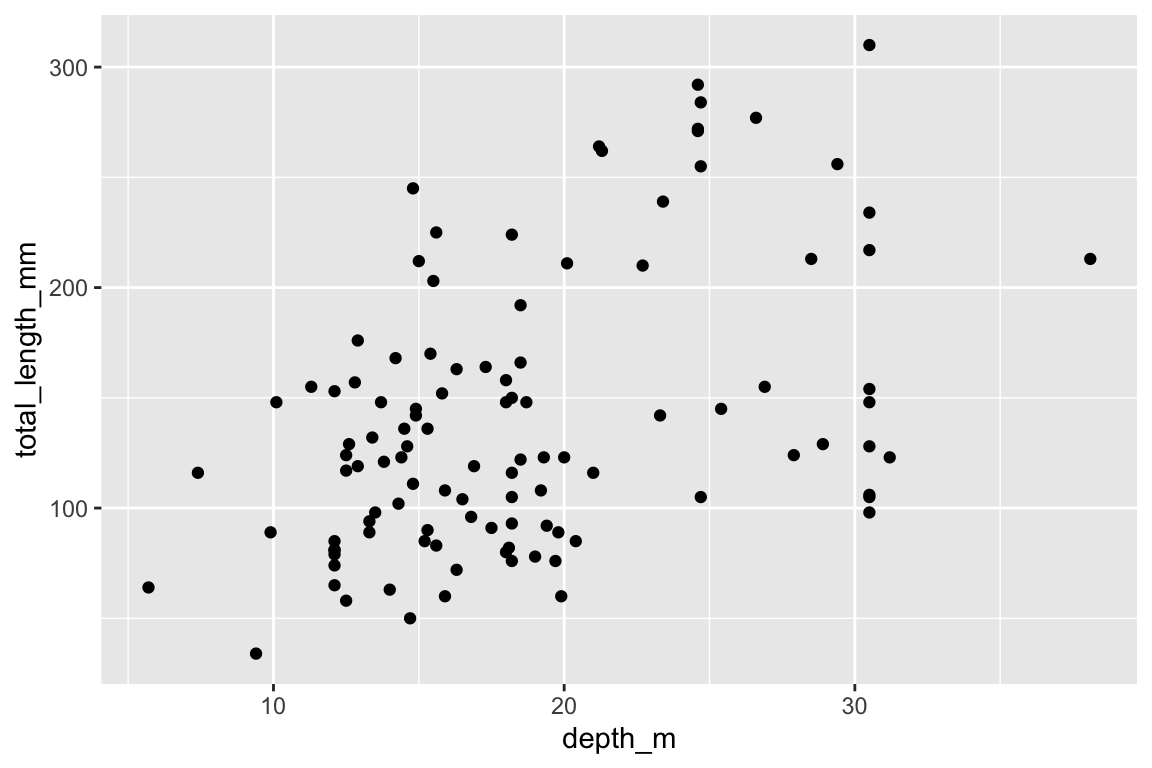
3. Specify the geometric Representation
4. Modify Geoms as Needed
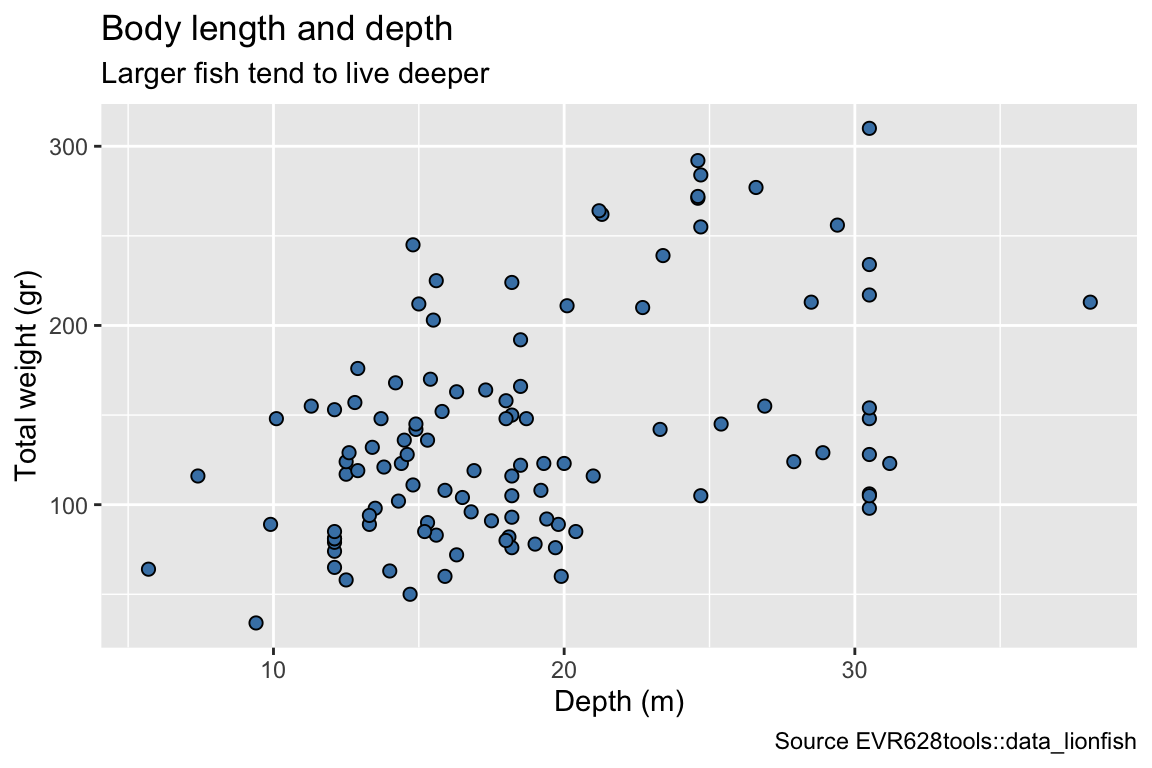
5. Modify Labels as Needed
5. Modify Labels as Needed
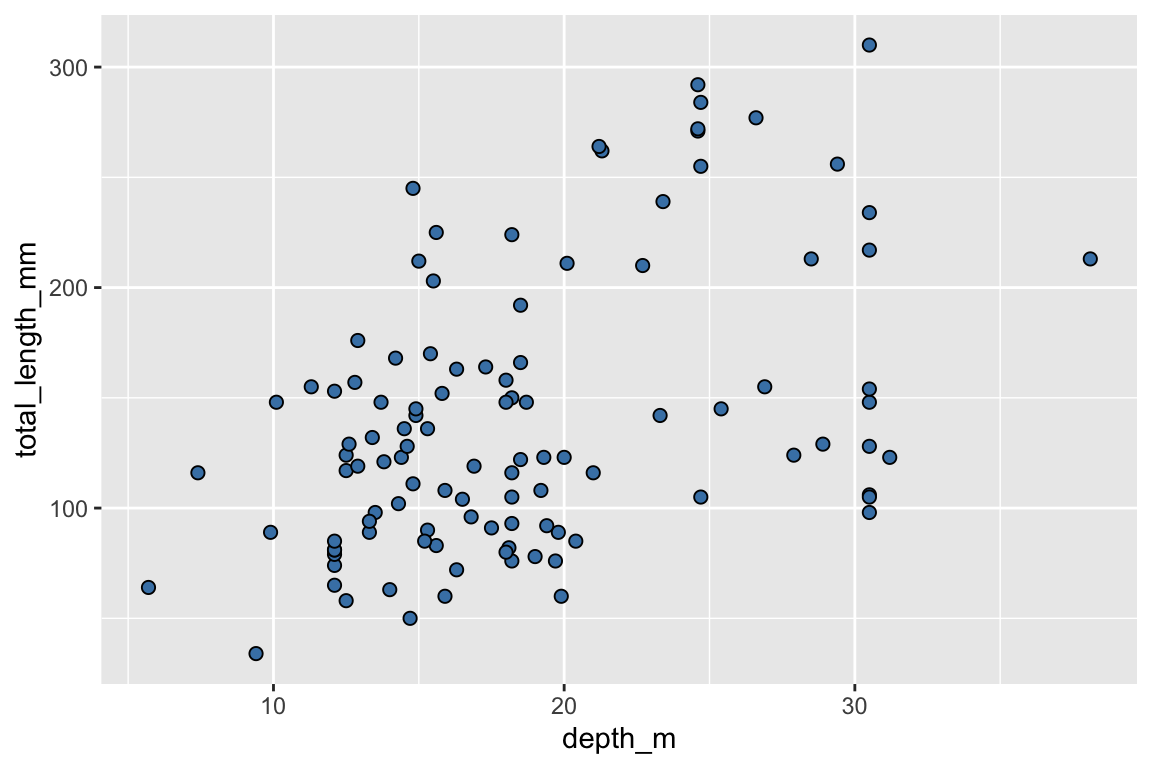
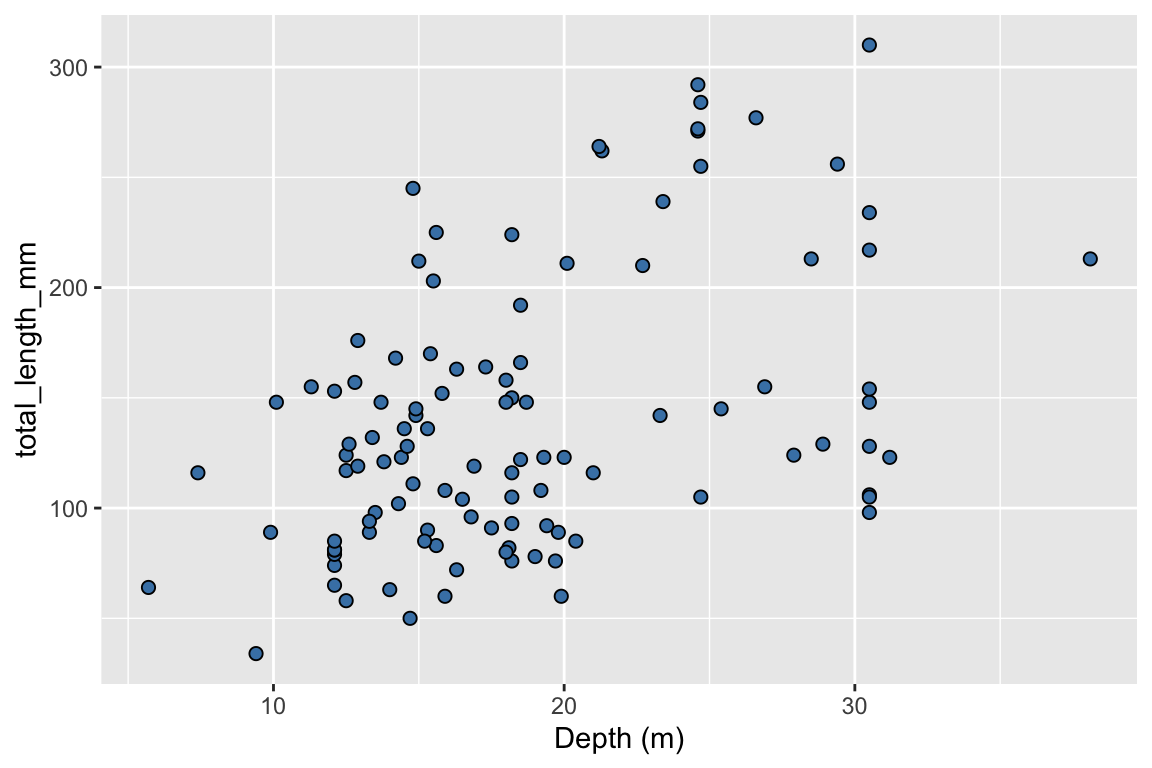
ggplot(data = data_lionfish,
mapping = aes(x = depth_m, y = total_length_mm)) +
geom_point(shape = 21, fill = "steelblue", size = 2) +
labs(x = "Depth (m)",
y = "Total length (mm)",
title = "Body length and depth",
subtitle = "Larger fish tend to live deeper",
caption = "Source EVR628tools::data_lionfish")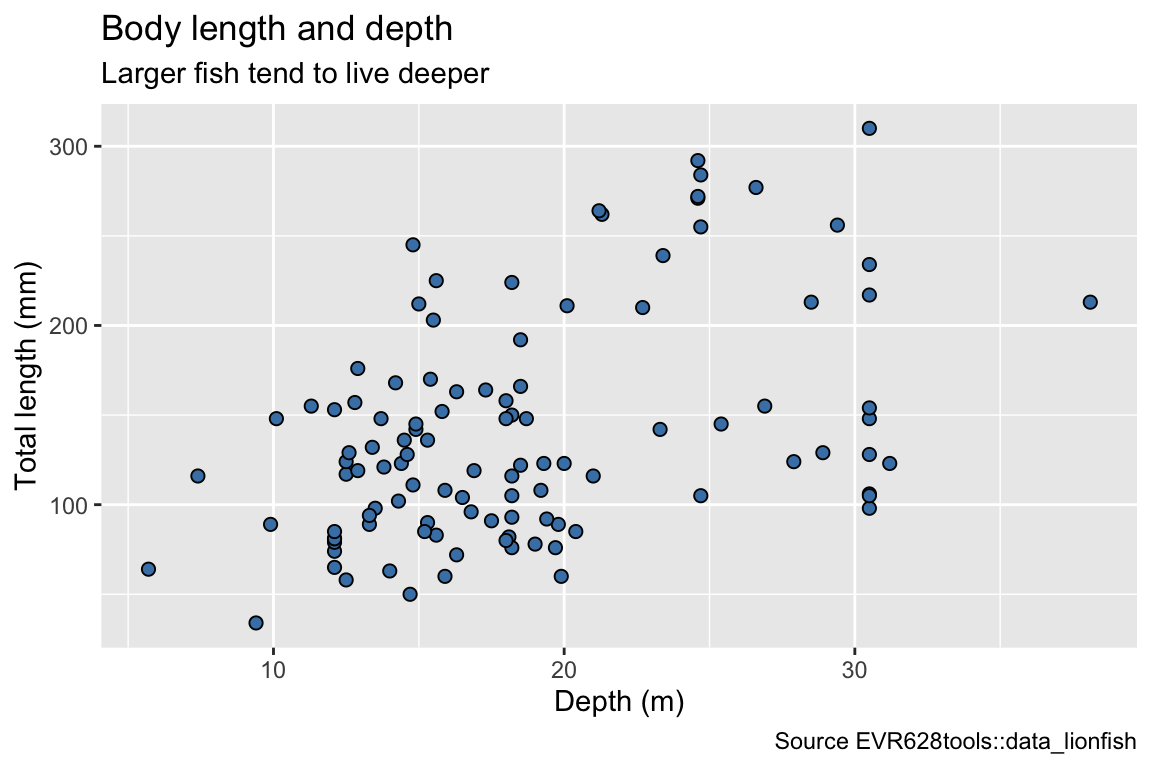
Resources
ggplot2cheatsheet- Package vignettes
- Wickham (2010)
Let’s get coding
Exercises
My guide for live coding