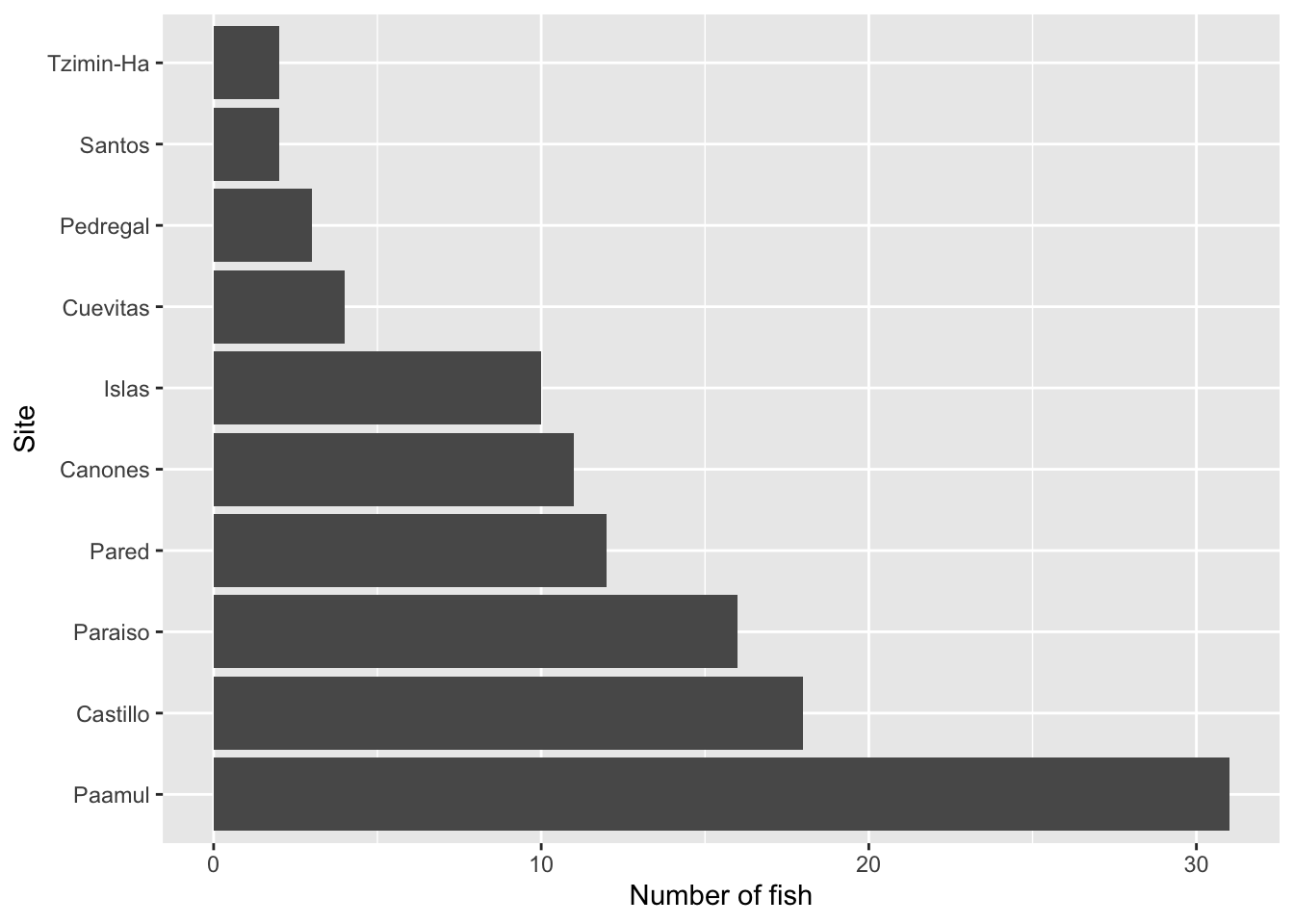
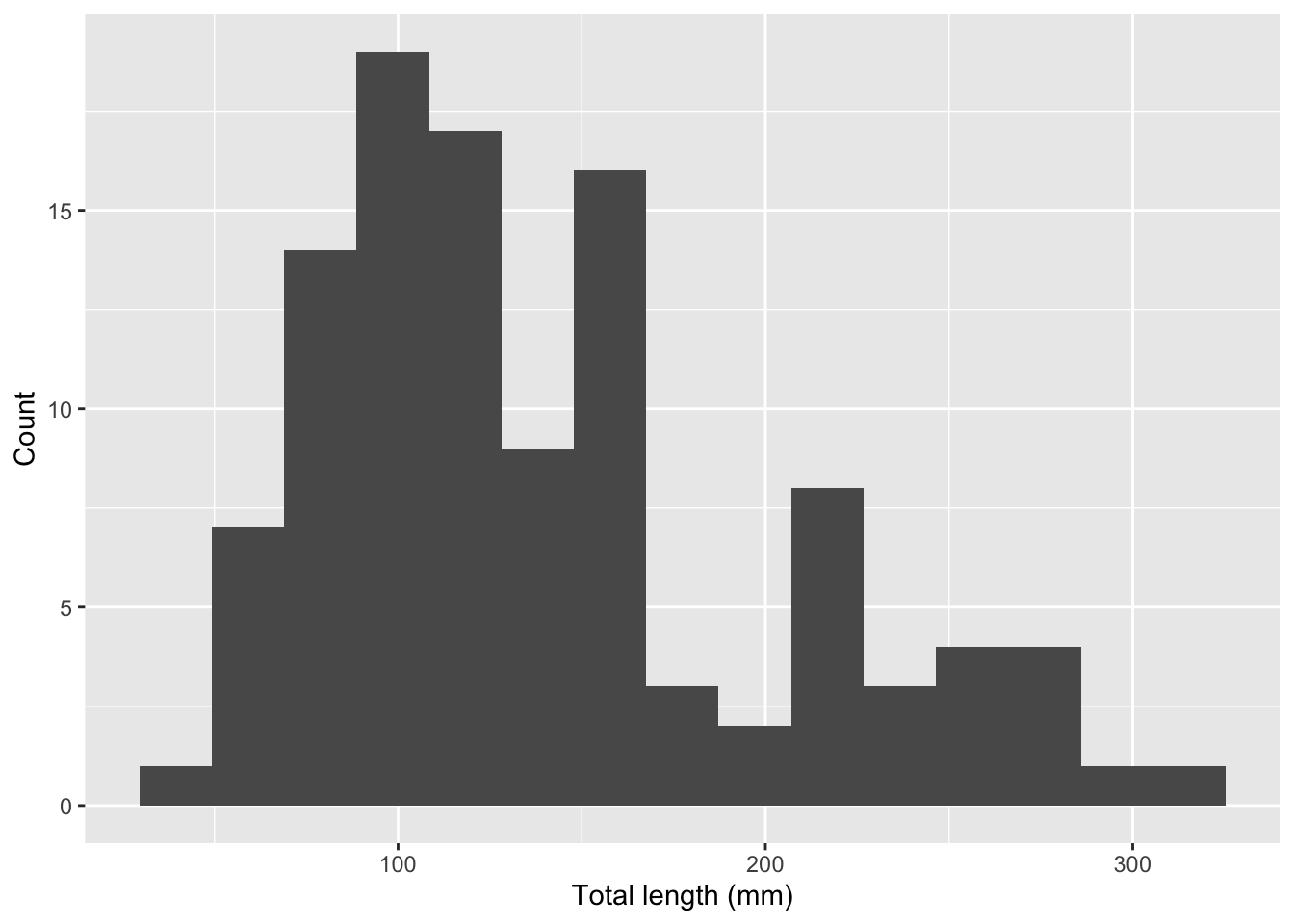
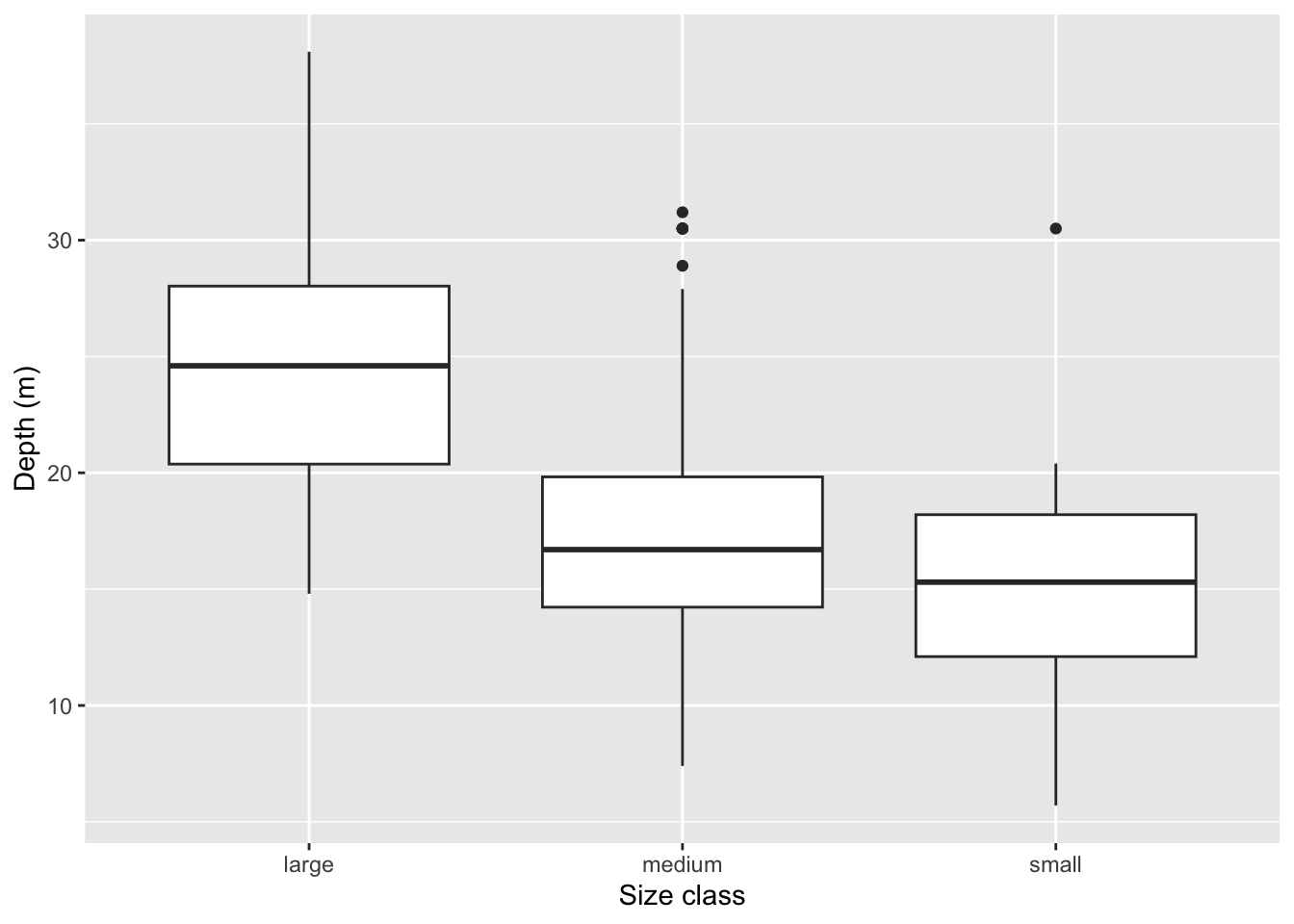
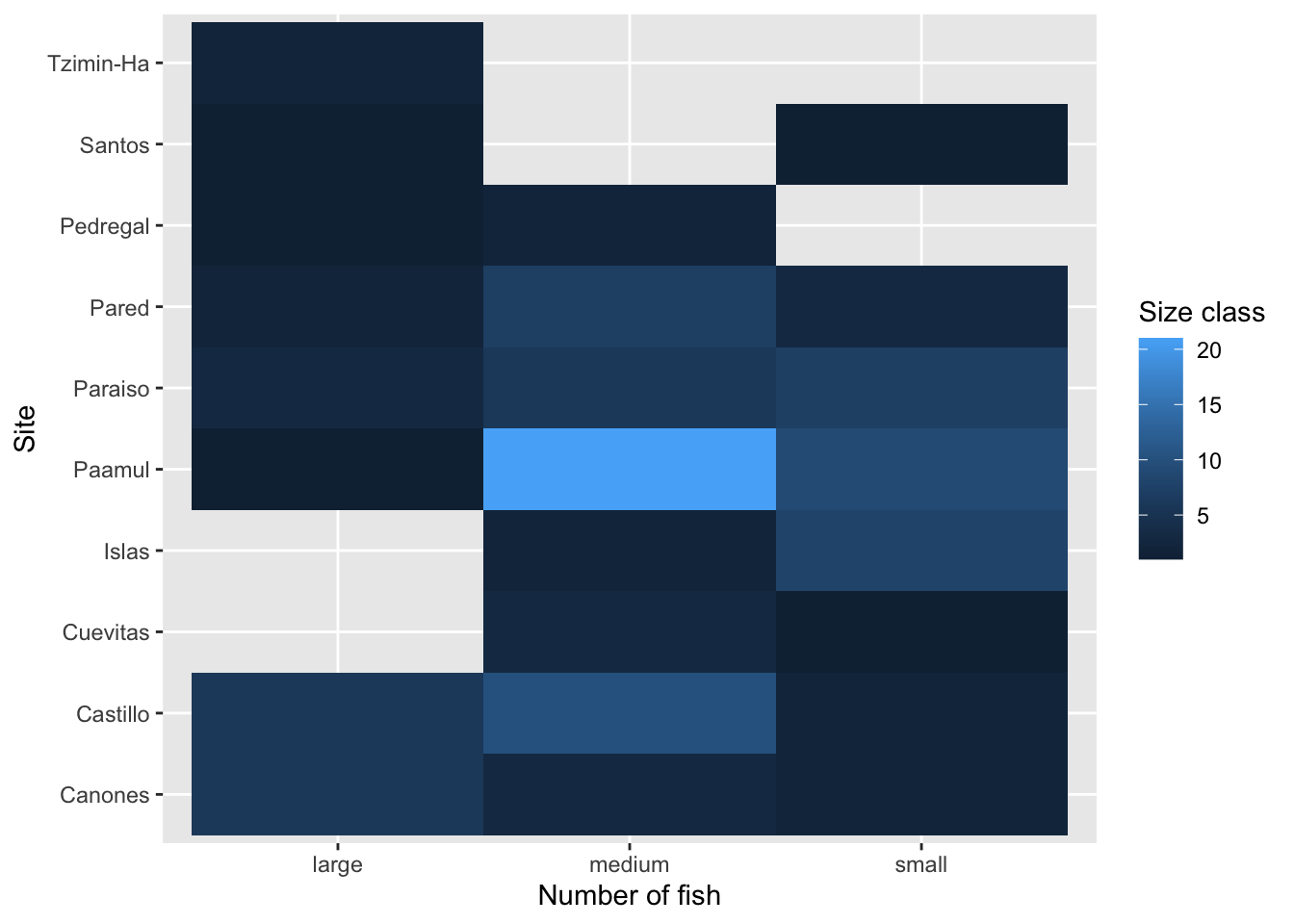
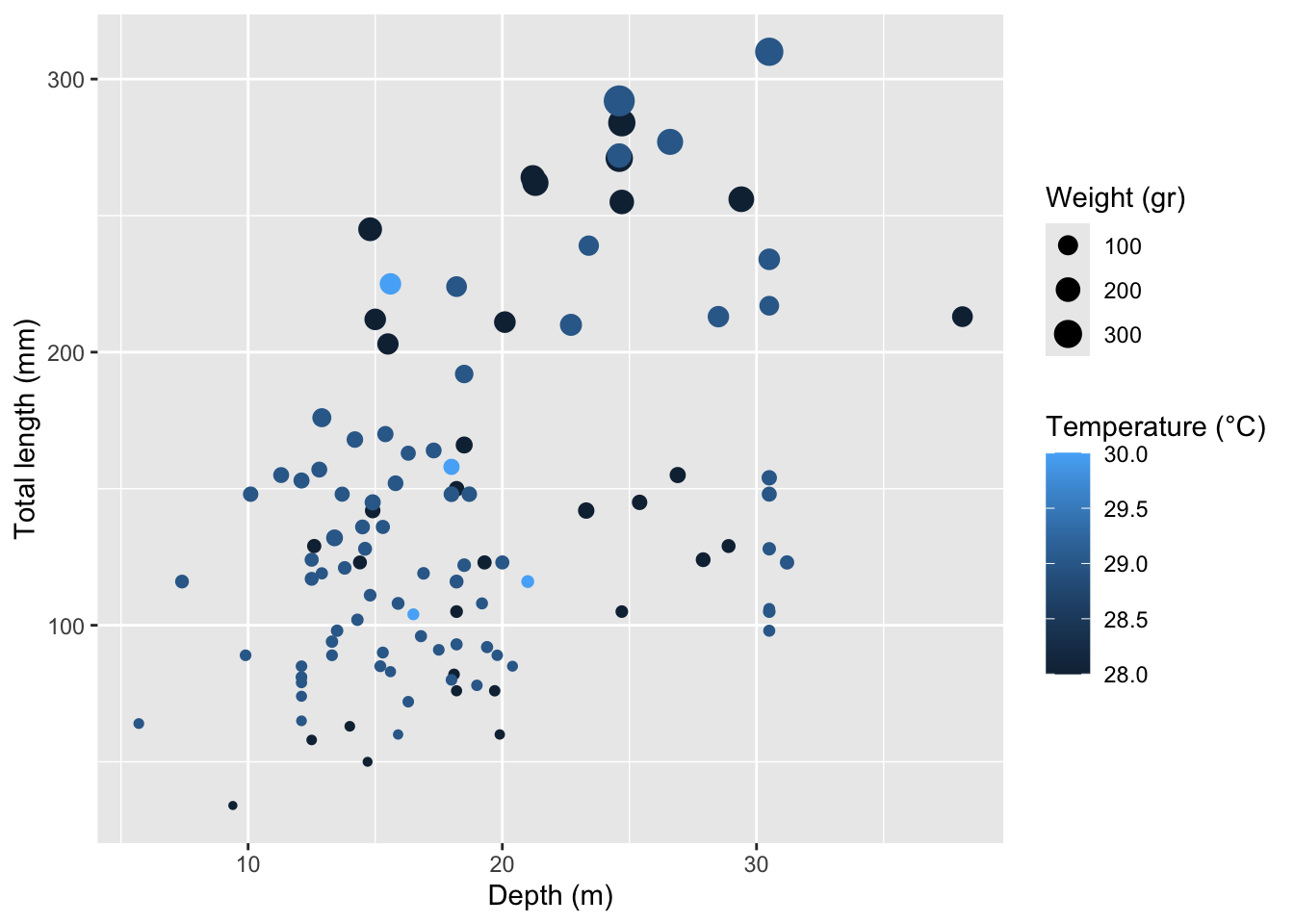
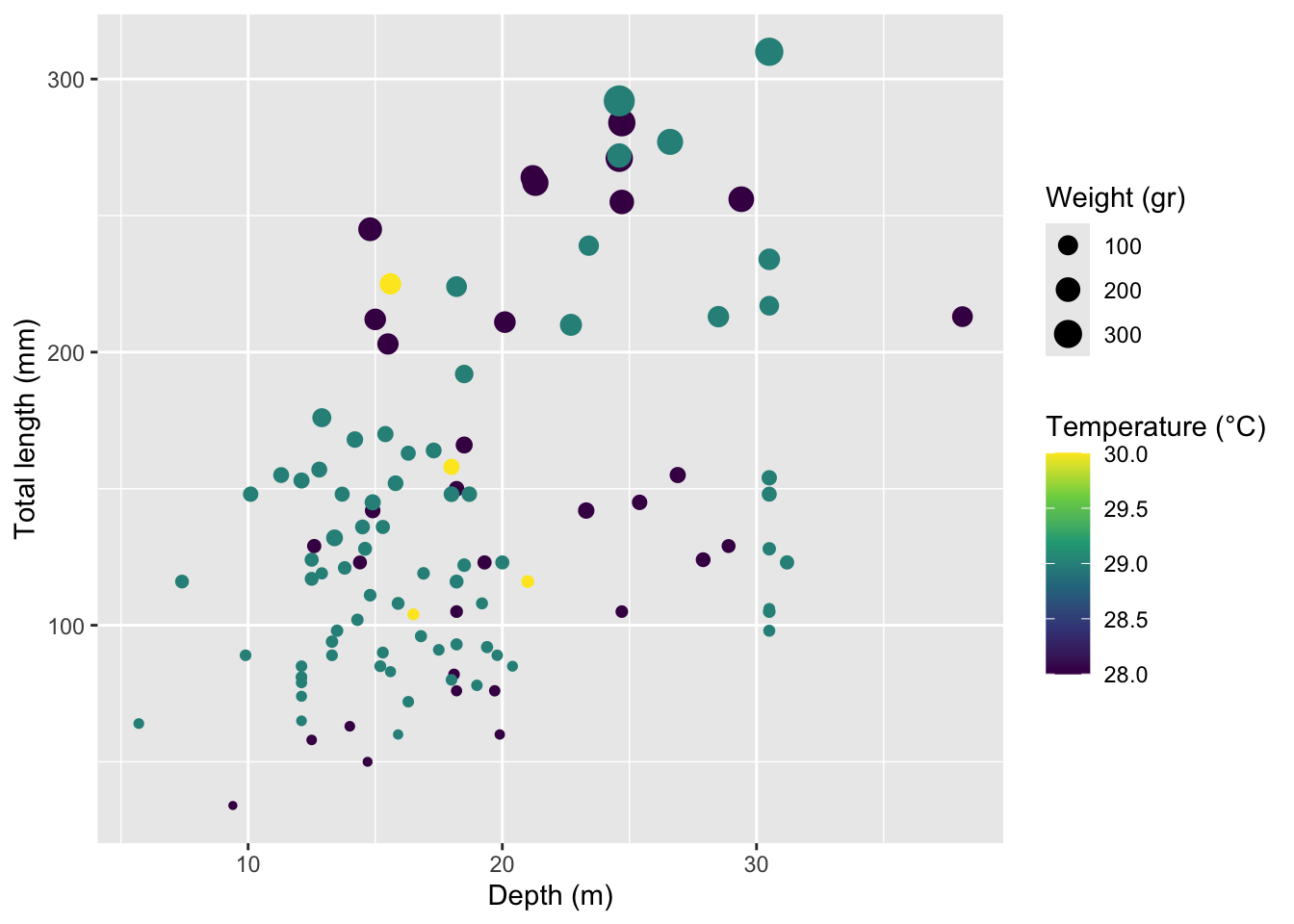
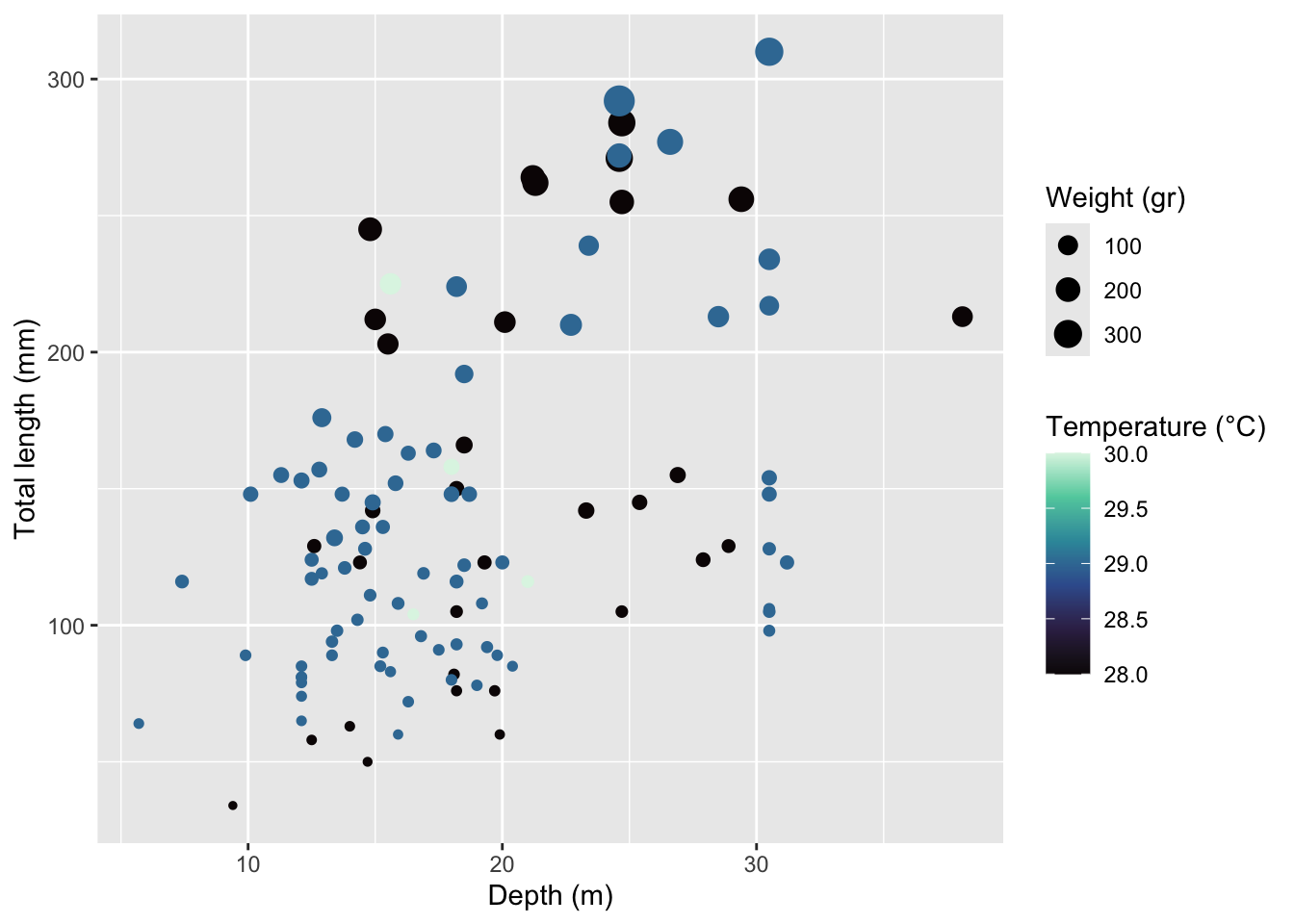
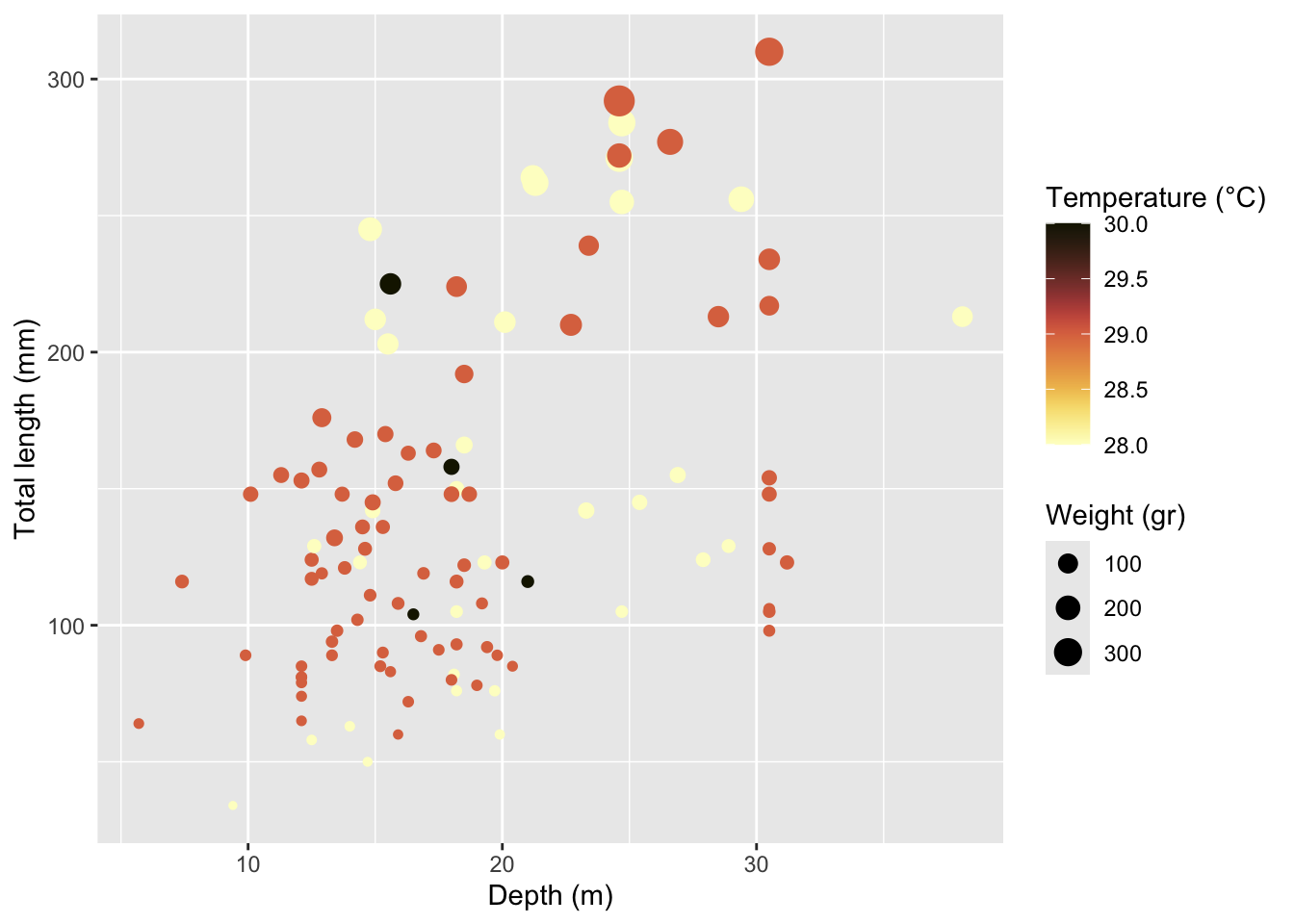
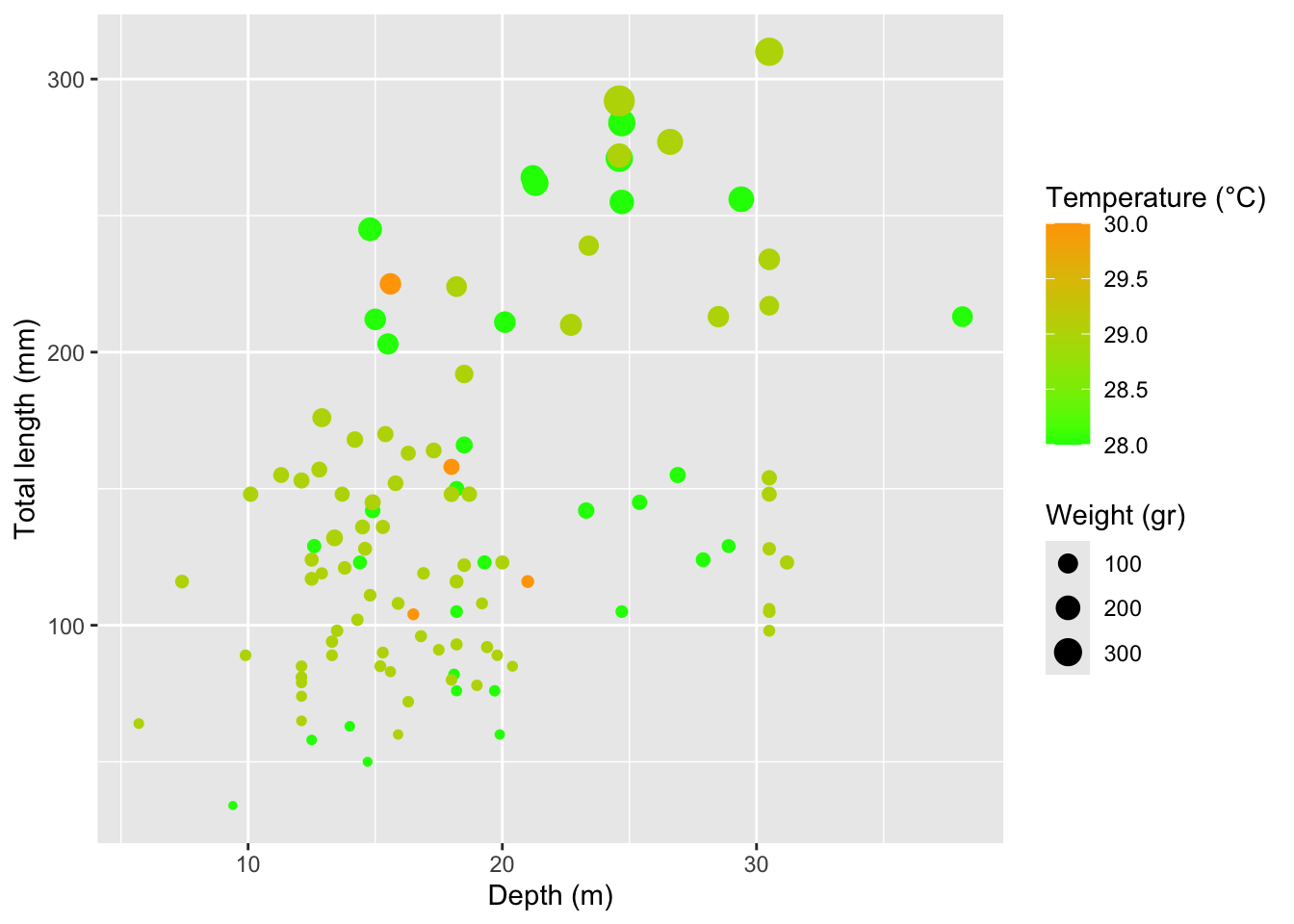
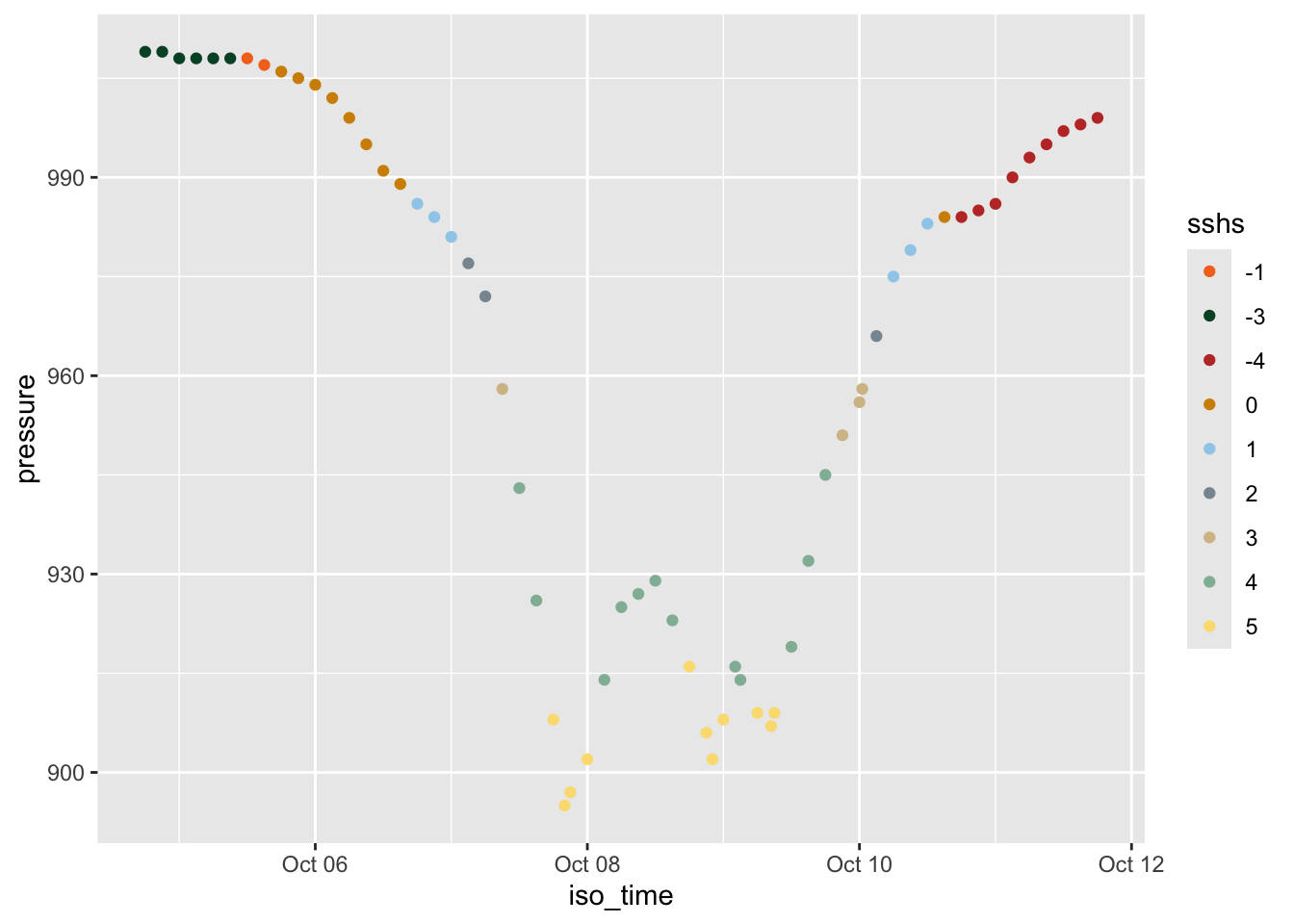
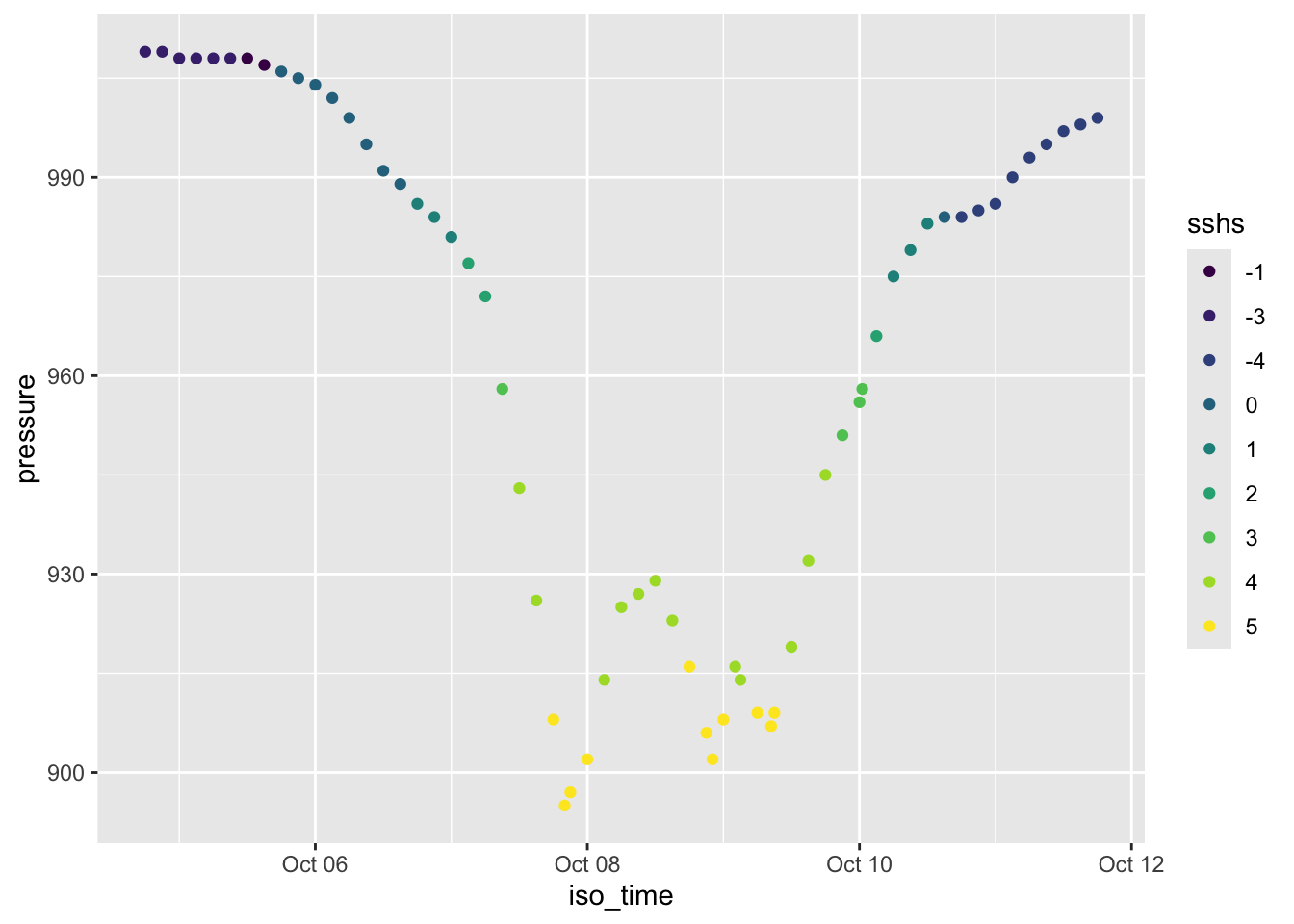
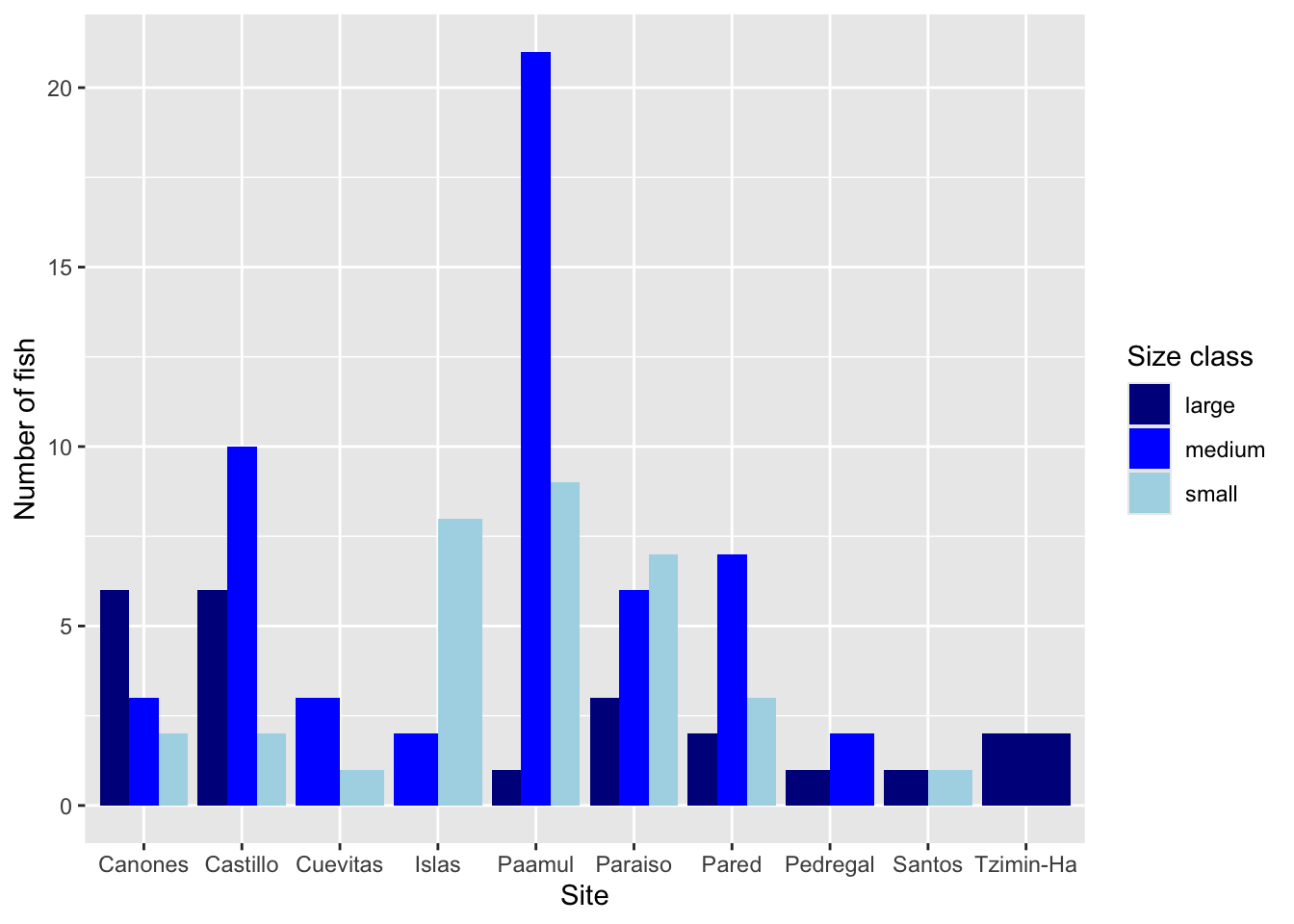
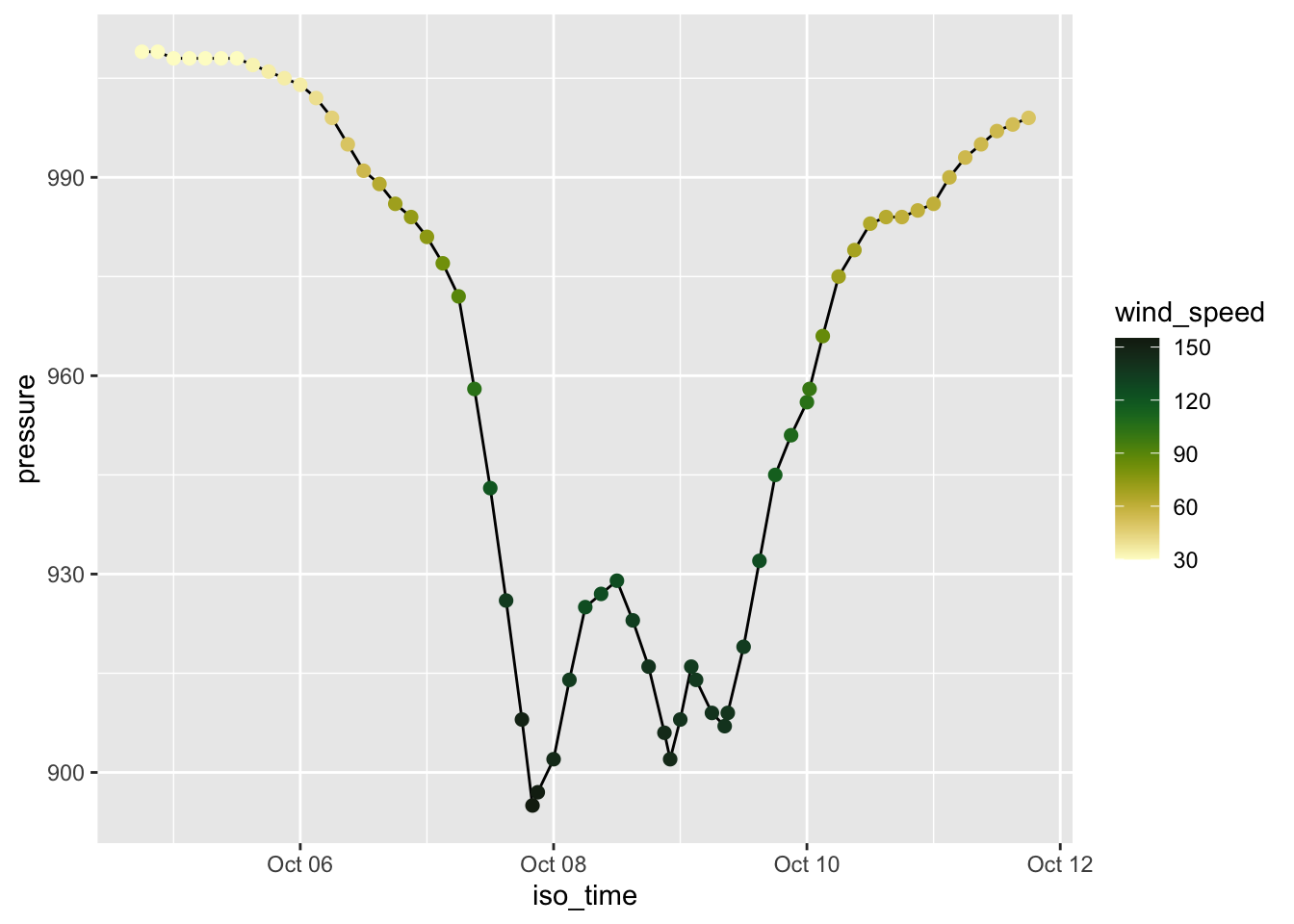
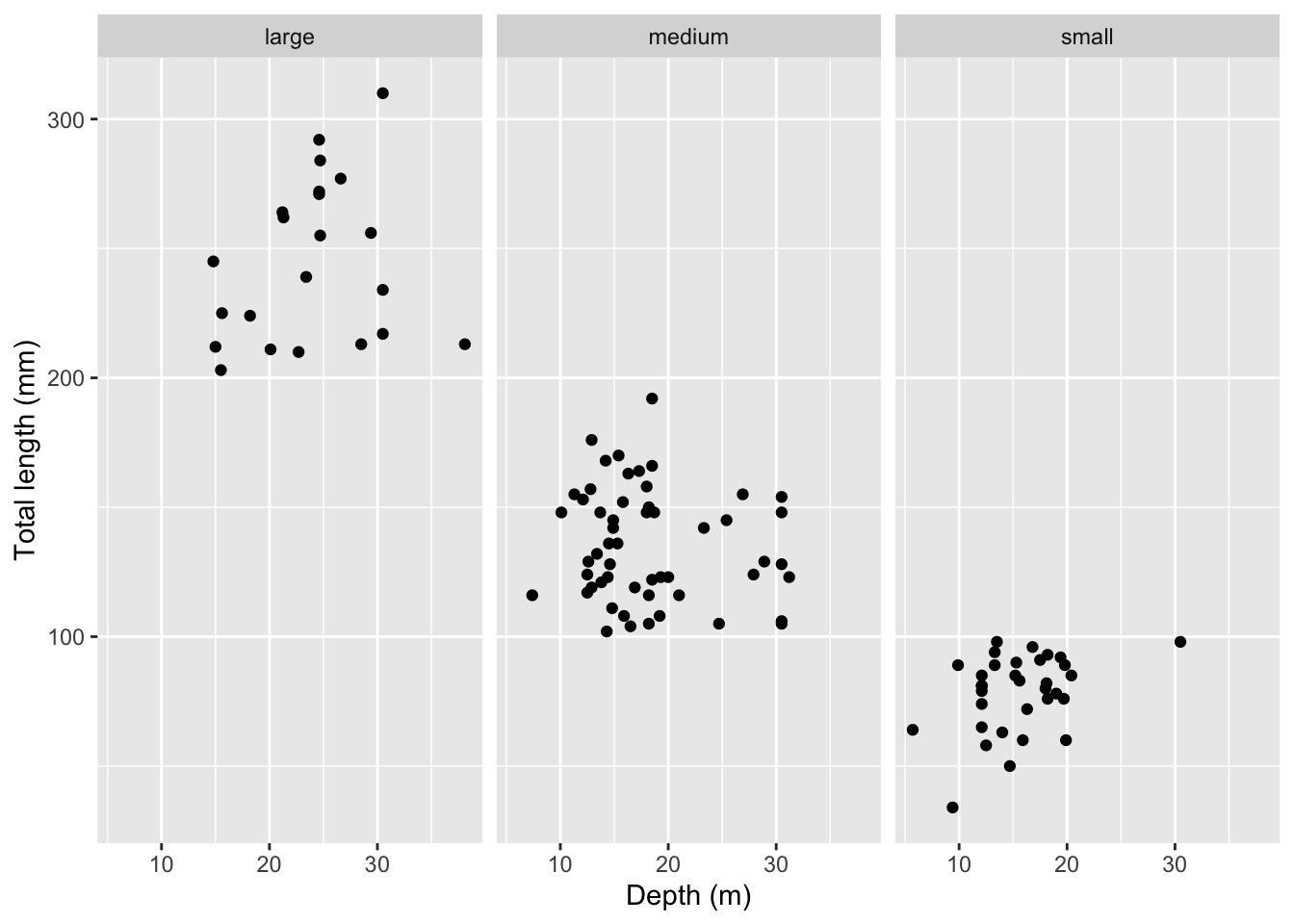
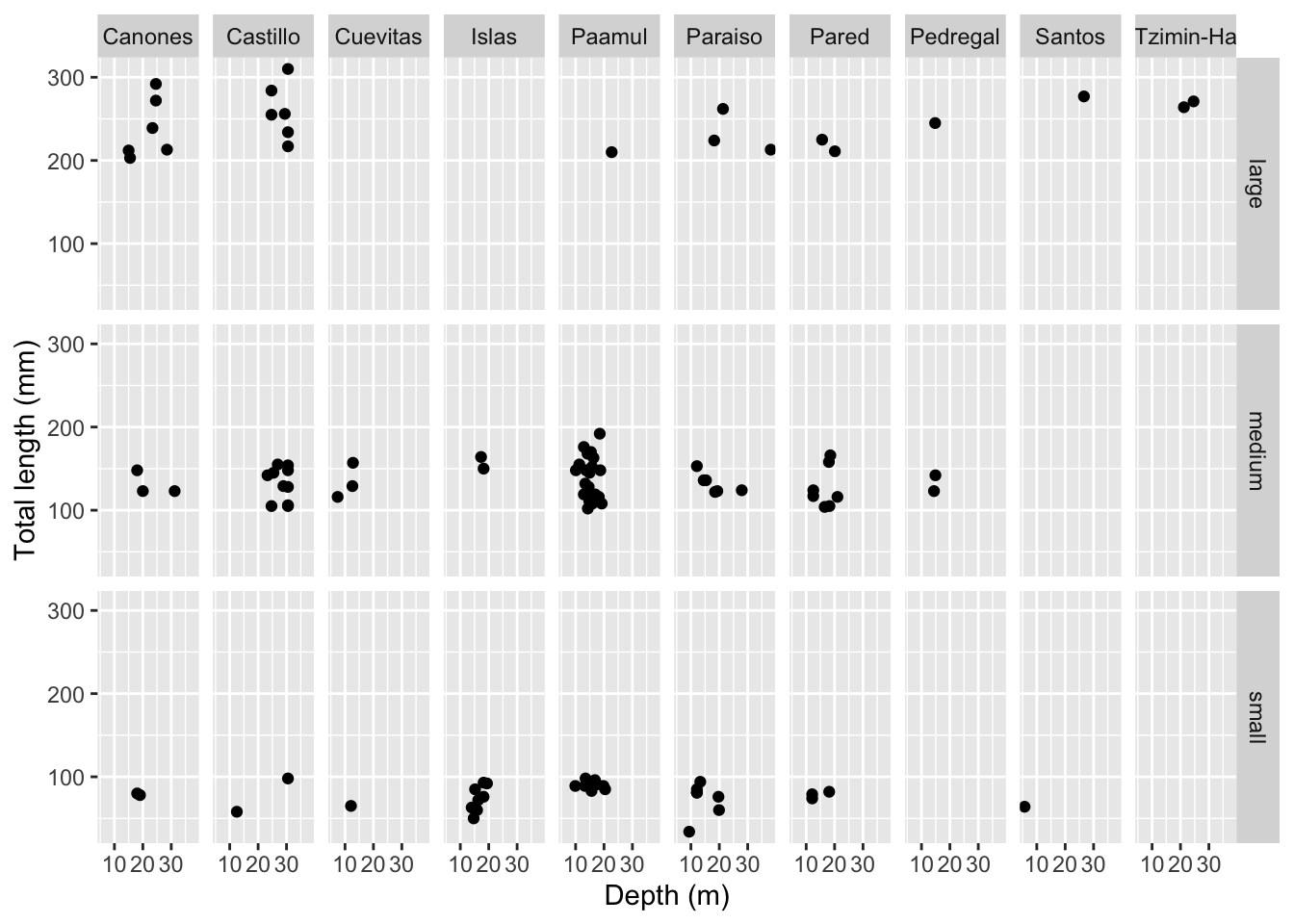
Rows: 109
Columns: 9
$ id <chr> "001-Po-16/05/10", "002-Po-29/05/10", "003-Pd-29/05/10…
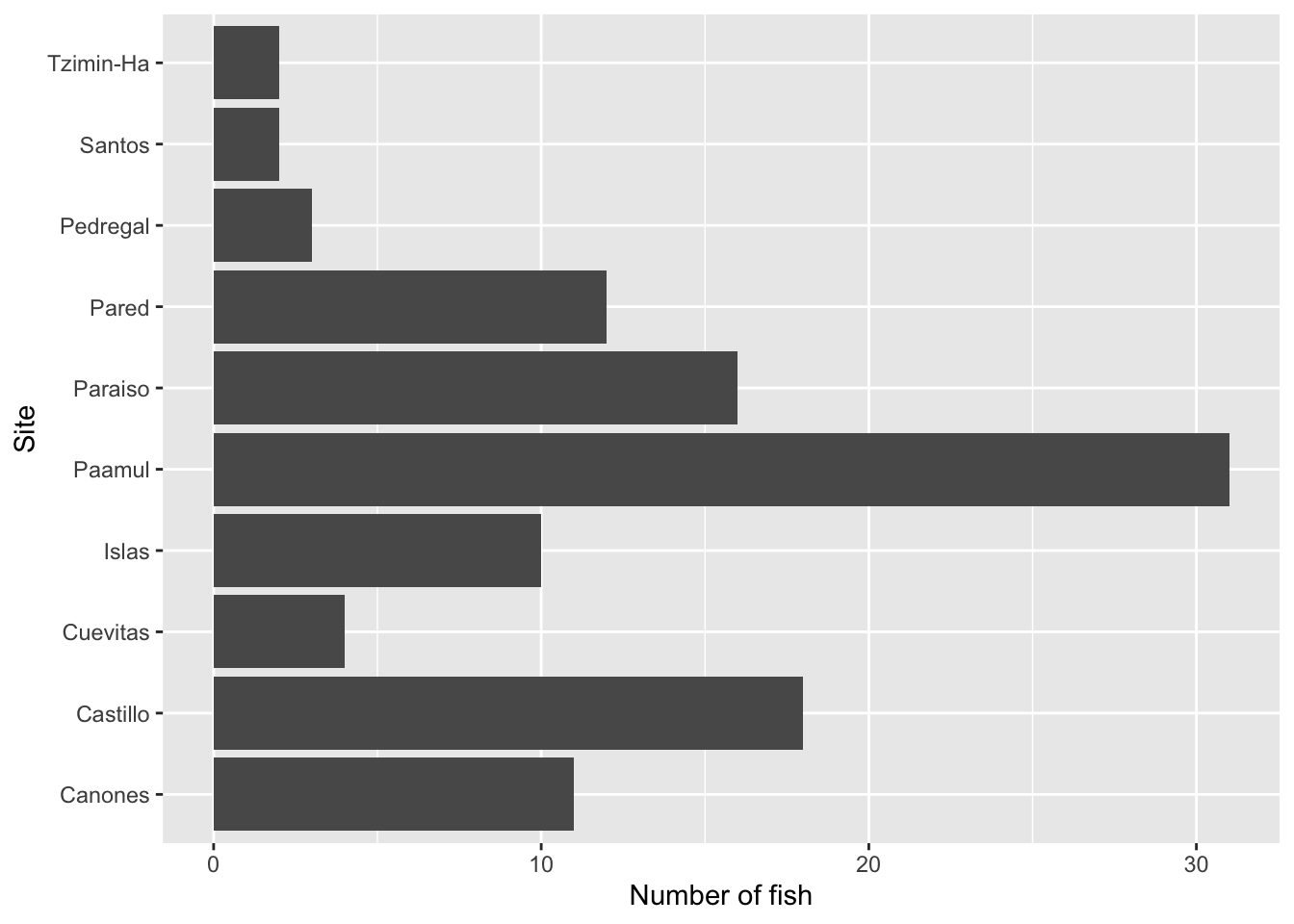
$ site <chr> "Paraiso", "Paraiso", "Pared", "Canones", "Canones", "…
$ lat <dbl> 20.48361, 20.48361, 20.50167, 20.47694, 20.47694, 20.5…
$ lon <dbl> -87.22611, -87.22611, -87.21167, -87.23278, -87.23278,…
$ total_length_mm <dbl> 213, 124, 166, 203, 212, 210, 132, 122, 224, 117, 211,…
$ total_weight_gr <dbl> 112.70, 27.60, 52.30, 123.10, 129.00, 138.75, 50.29, 1…
$ size_class <chr> "large", "medium", "medium", "large", "large", "large"…
$ depth_m <dbl> 38.1, 27.9, 18.5, 15.5, 15.0, 22.7, 13.4, 18.5, 18.2, …
$ temperature_C <dbl> 28, 28, 28, 28, 28, 29, 29, 29, 29, 29, 28, 28, 28, 28…