Scaling up your data visualizations (Part 1)
EVR 628- Intro to Environmental Data Science
Juan Carlos Villaseñor-Derbez (JC)
Rosenstiel School of Marine, Atmospheric, and Earth Science and Institute for Data Science and Computing
Learning Objectives
By the end of this week, you should be able to:
- Create complex figures
- Develop simple documents
This week
- Review of
dplyrandtidyr - Review of
ggplot() - Layering
geoms - Subplots with facets
ggplotextensionsggridgescowplot
- Position adjustments
- Labels
- Summarizing data on the fly
- Topics above not covered AND
- Quarto markdown
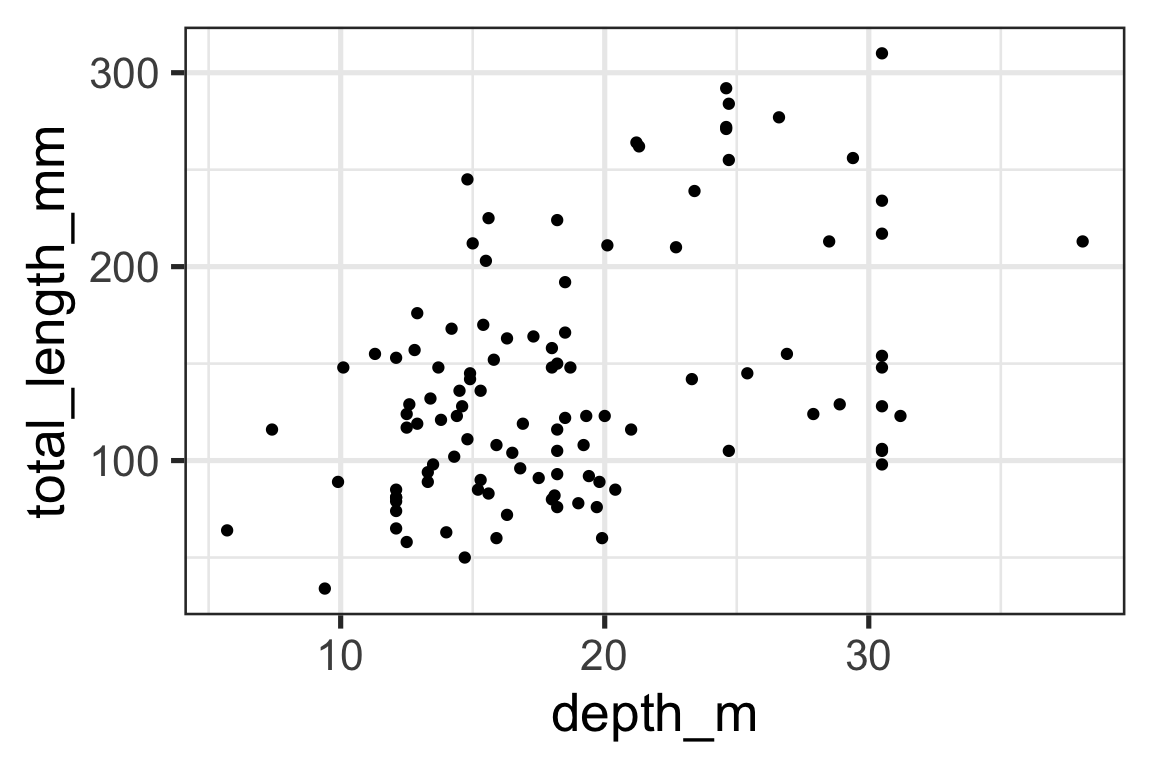
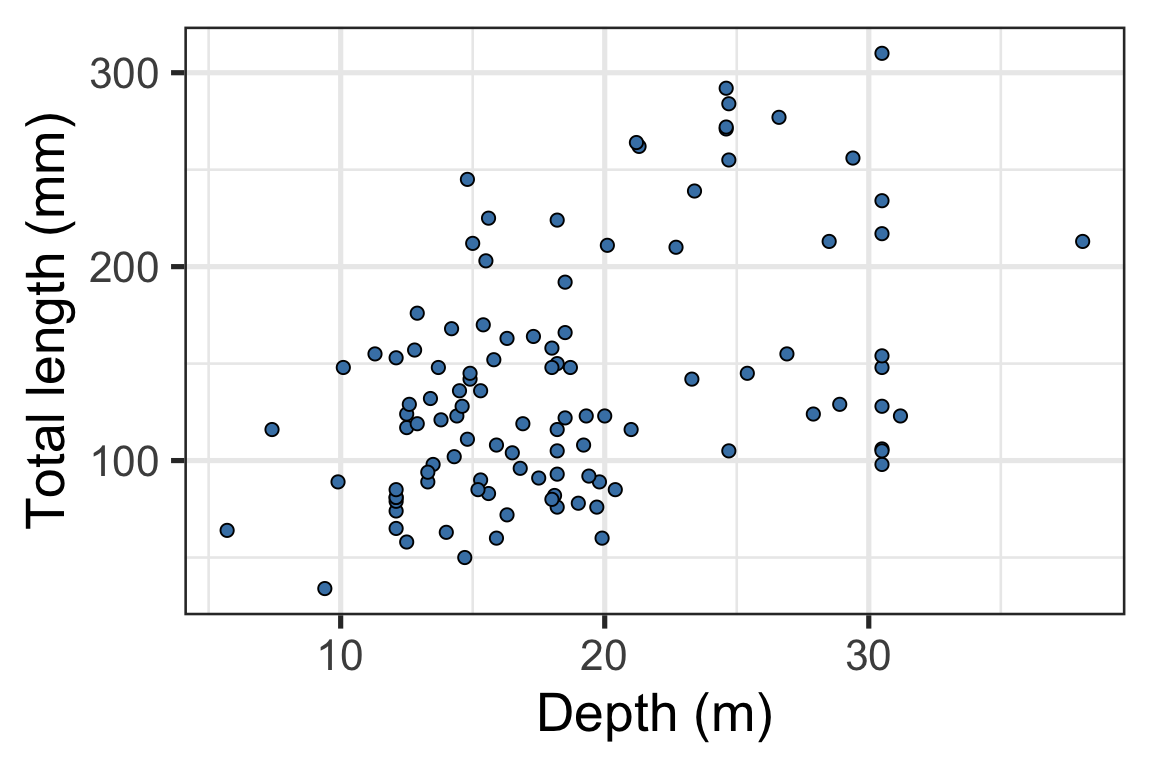
Review of ggplot()
Review of ggplot()
Build a plot with three steps
- Specify your data in
ggplot() - Specify your x (and y) axis
aesthetic mappings inaes() - Specify your
geometric representation withgeom_*
And maybe:
- Modify
geoms as needed - Modify your labels as needed
1. Specify the Data
2. Specify the aesthetics
3. Specify the geometric Representation
4-5. Modify geoms and labels
Layering geoms
Layering geoms
- The
groupaesthetic - non-data geoms
- smooth geom
- geom-level vs global-level aesthetics
geoms on top of geoms 1
Code
ggplot(data = data_lionfish,
mapping = aes(x = total_length_mm, y = total_weight_gr)) +
geom_smooth(color = "black", linetype = "dashed") +
geom_vline(xintercept = c(100, 200), linetype = "dashed") +
geom_point(aes(color = size_class), size = 2) +
scale_color_manual(values = palette_UM(3)) +
labs(x = "Total length (mm)",
y = "Total weight (gr)",
color = "Size class") +
theme(legend.position = "inside",
legend.justification = c(0, 1),
legend.position.inside = c(0, 1),
legend.background = element_blank())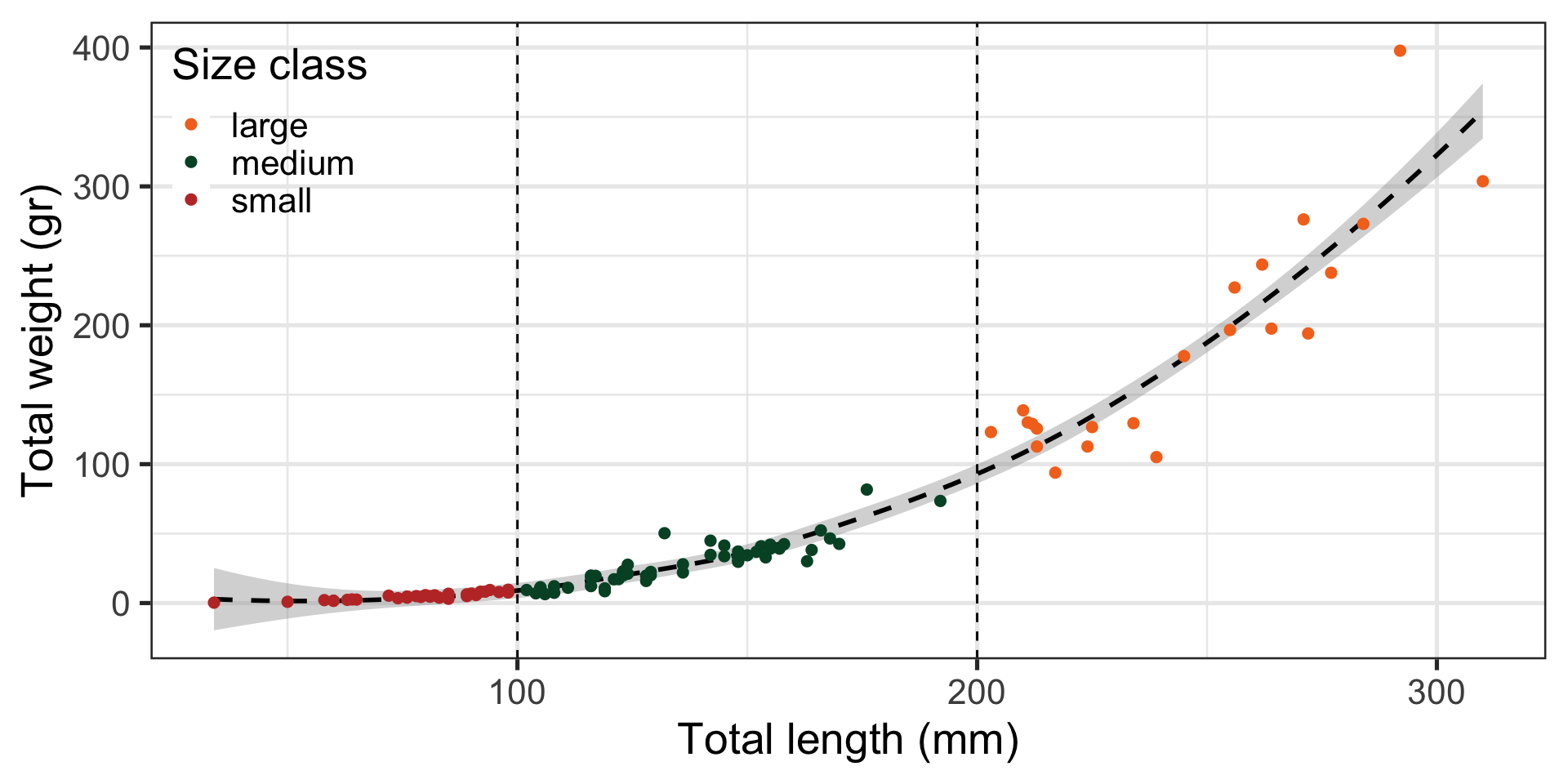
geoms on top of geoms 2
geoms on top of geoms 2
geoms on top of geoms 2
geoms on top of geoms 3
Code
data(data_mhw_events)
ggplot(data = data_mhw_events,
mapping = aes(x = date_peak, y = intensity_max,
color = intensity_max)) +
geom_linerange(mapping = aes(ymin = 0,
ymax = intensity_max),
linewidth = 1) +
geom_point(size = 2) +
scale_color_gradient(low = "gray90", high = "red") +
labs(x = "Date peak",
y = "MHW Intensity (°C)",
color = "MHW Intensity (°C days)") +
theme(legend.position = "bottom",
legend.title.position = "top",
legend.key.width = unit(1, "cm"))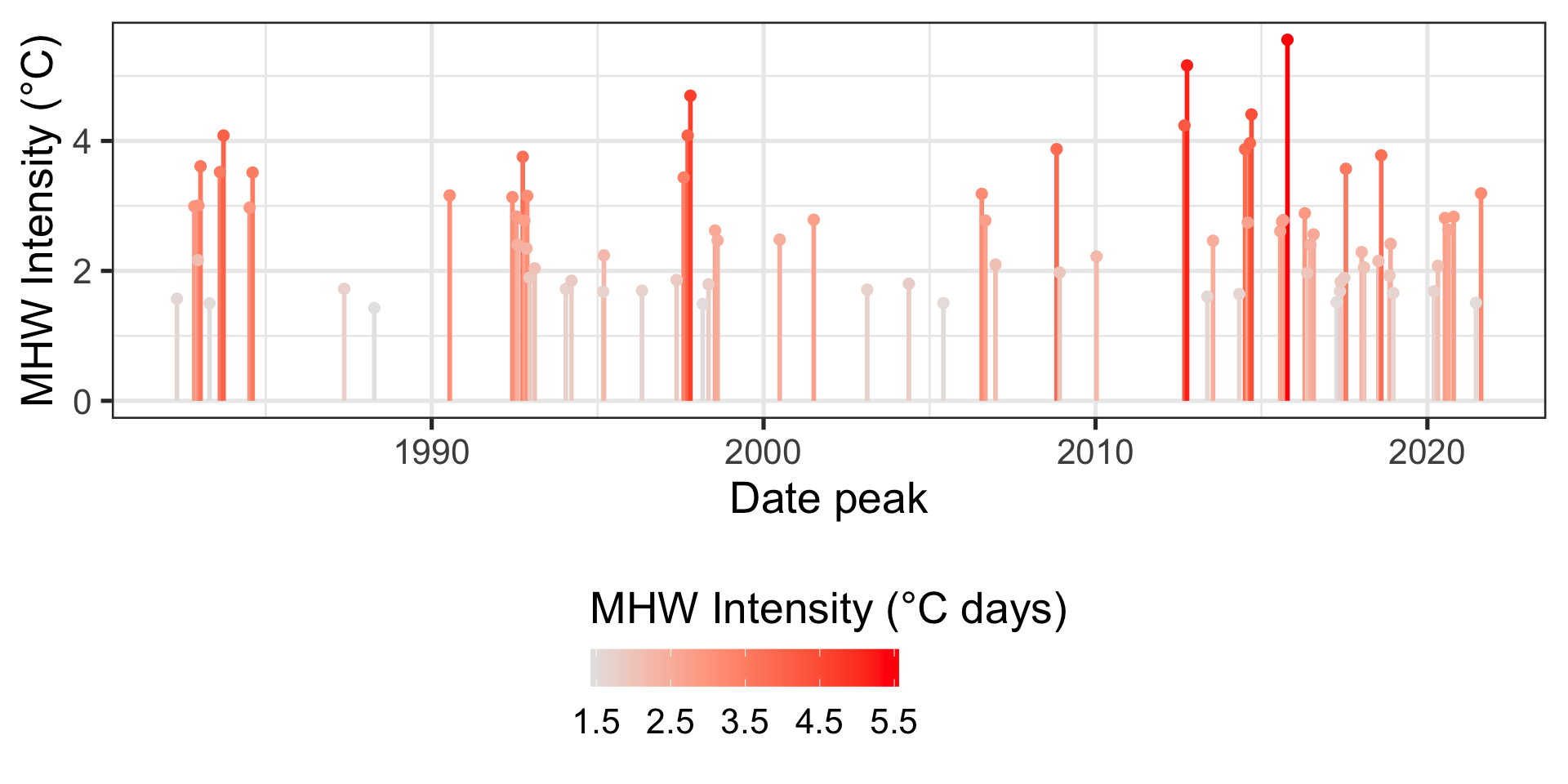
Facetting
Facetting with wrap
When specifying groups and colors is not enough
Facetting with grid
Code
tidy_kelp <- data_kelp |>
filter(genus_species %in% c("Embiotoca jacksoni",
"Embiotoca lateralis"),
location %in% c("ASA", "ERE", "ERO")) |>
pivot_longer(cols = starts_with("TL_"),
names_to = "total_length",
values_to = "N",
values_drop_na = T) |>
group_by(location, site, transect, genus_species) |>
summarize(total_N = sum(N)) |>
group_by(location, site, genus_species) |>
summarize(mean_N = mean(total_N))
ggplot(data = tidy_kelp,
mapping = aes(x = site, y = mean_N)) +
geom_col() +
facet_grid(location ~ genus_species) +
labs(x = "Site", y = "Mean (org / tranect)")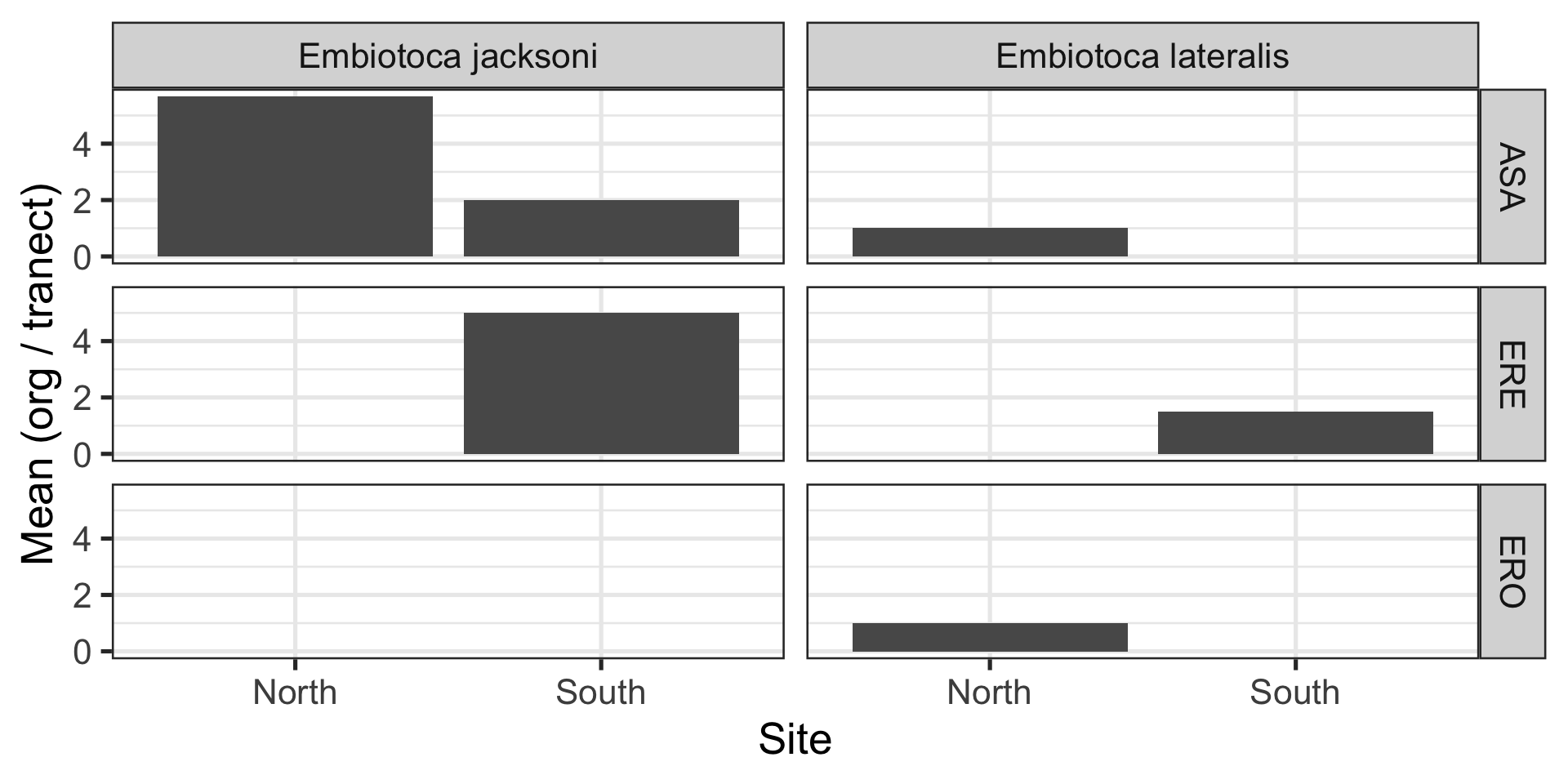
ggplot extensions
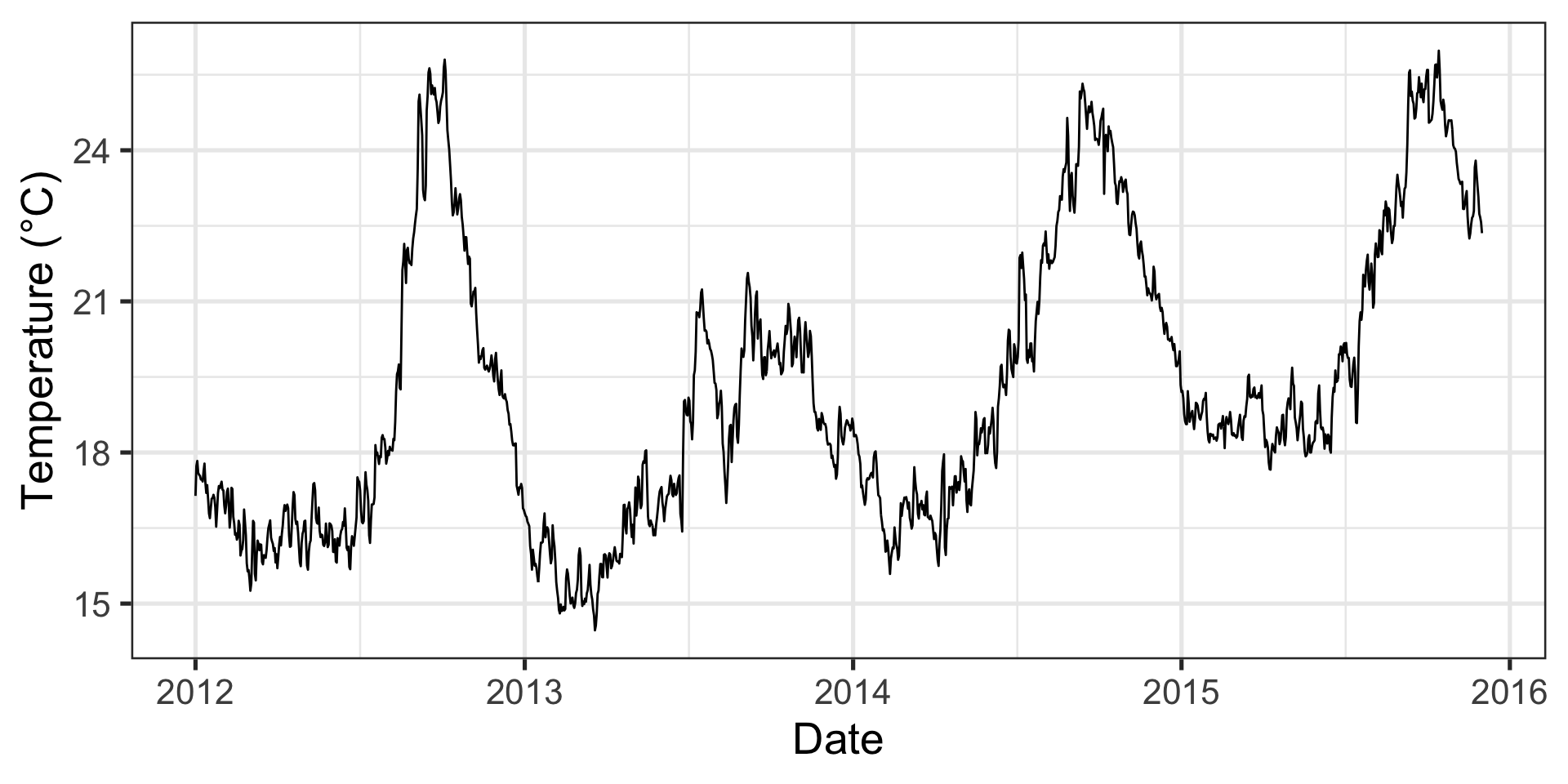
ggplot extension 1: cowplot
Code
library(cowplot)
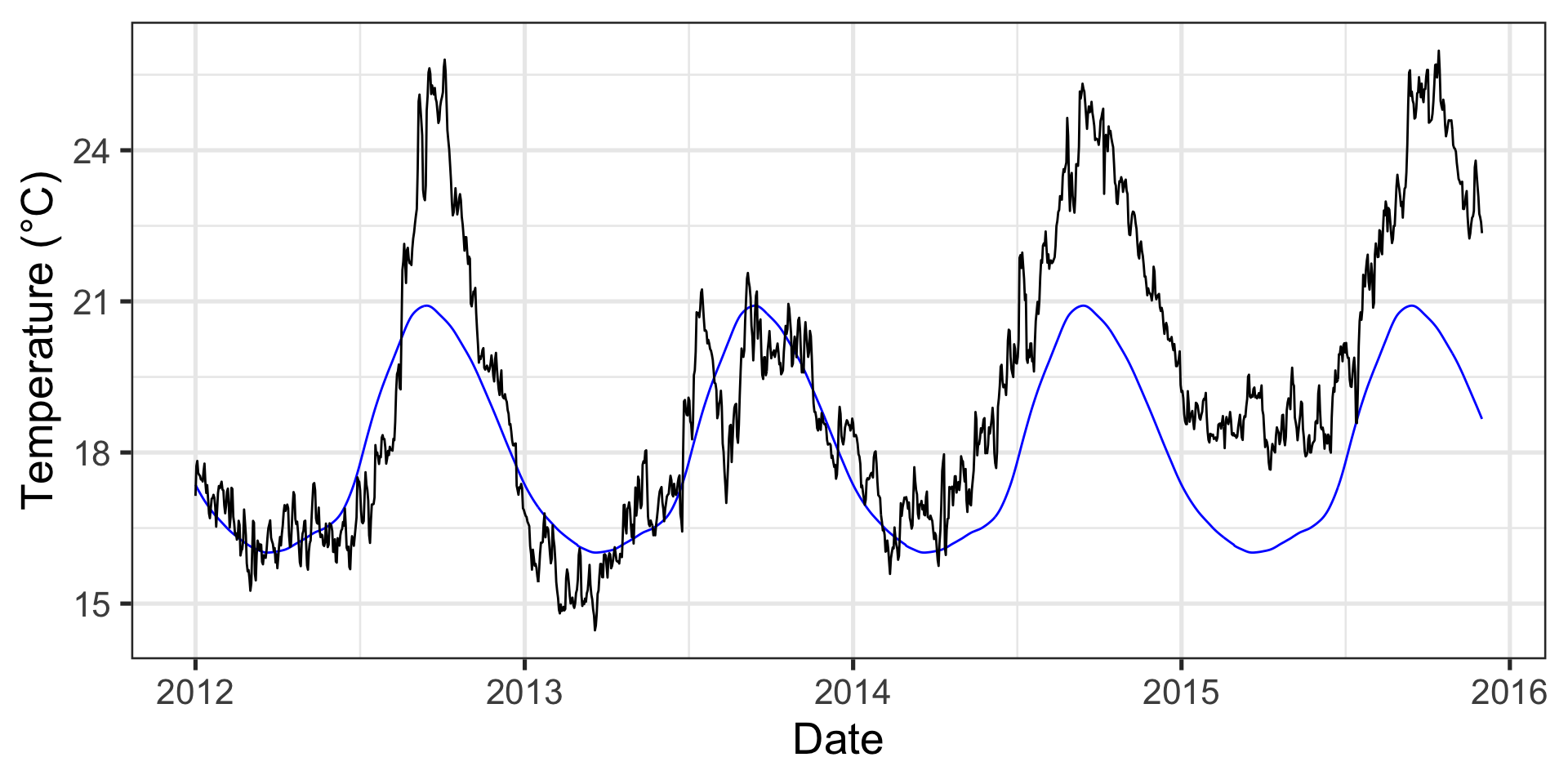
p1 <- ggplot(data = data_mhw_ts, aes(x = date, y = temp)) +
geom_line() +
geom_line(aes(y = seas), color = "blue") +
geom_line(aes(y = thresh), color = "red") +
labs(x = "Date", y = "Temperature (°C)")
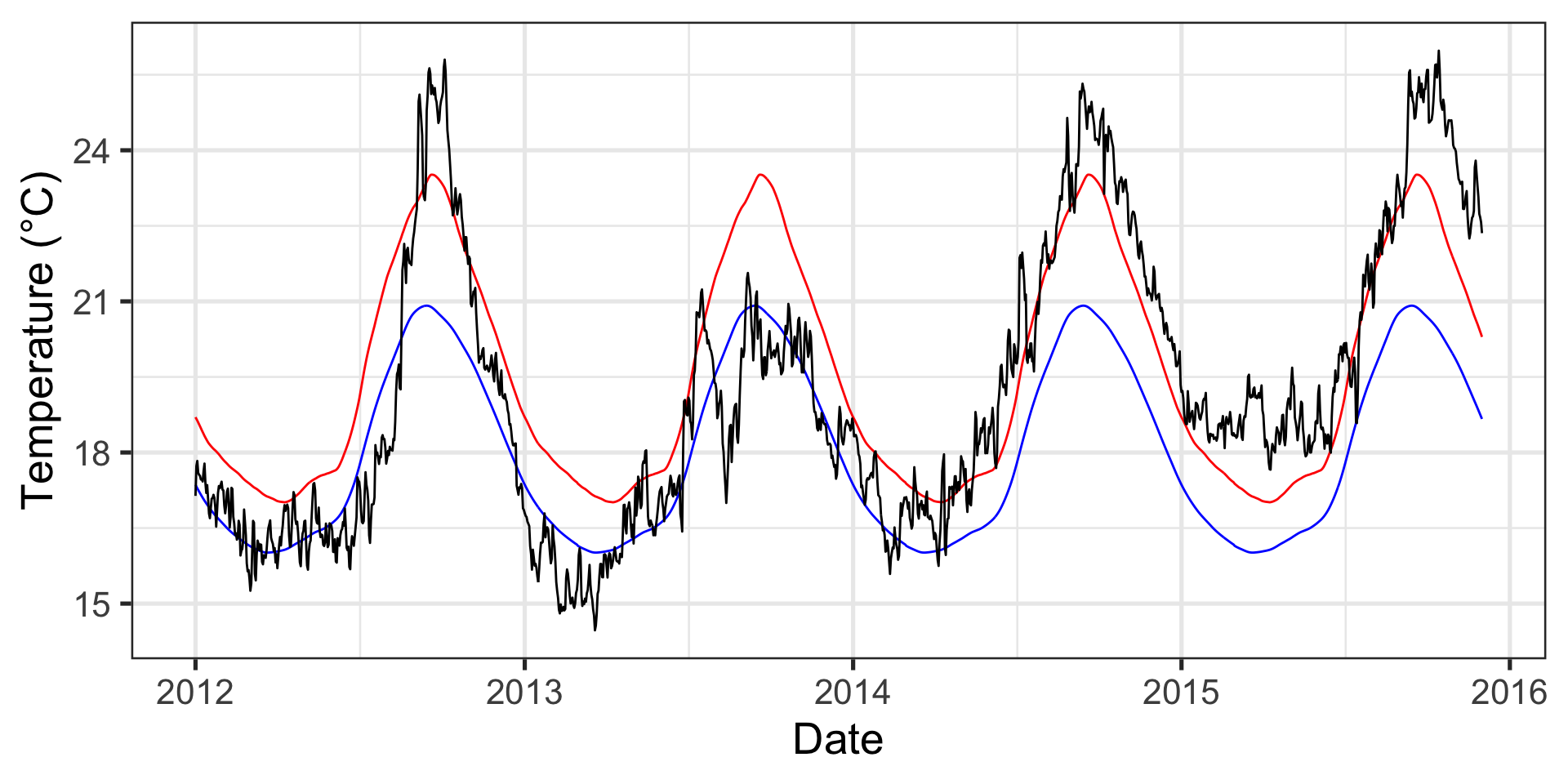
p2 <- ggplot(data = data_mhw_events,
mapping = aes(x = date_peak, y = intensity_max,
color = intensity_max)) +
geom_linerange(mapping = aes(ymin = 0,
ymax = intensity_max),
linewidth = 1) +
geom_point(size = 2) +
scale_color_gradient(low = "gray90", high = "red") +
labs(x = "Date peak",
y = "MHW Intensity (°C)",
color = "MHW Intensity (°C days)") +
theme(legend.position = "bottom",
legend.title.position = "top",
legend.key.width = unit(1, "cm"))
plot_grid(p1, p2, ncol = 1, rel_heights = c(0.5, 1))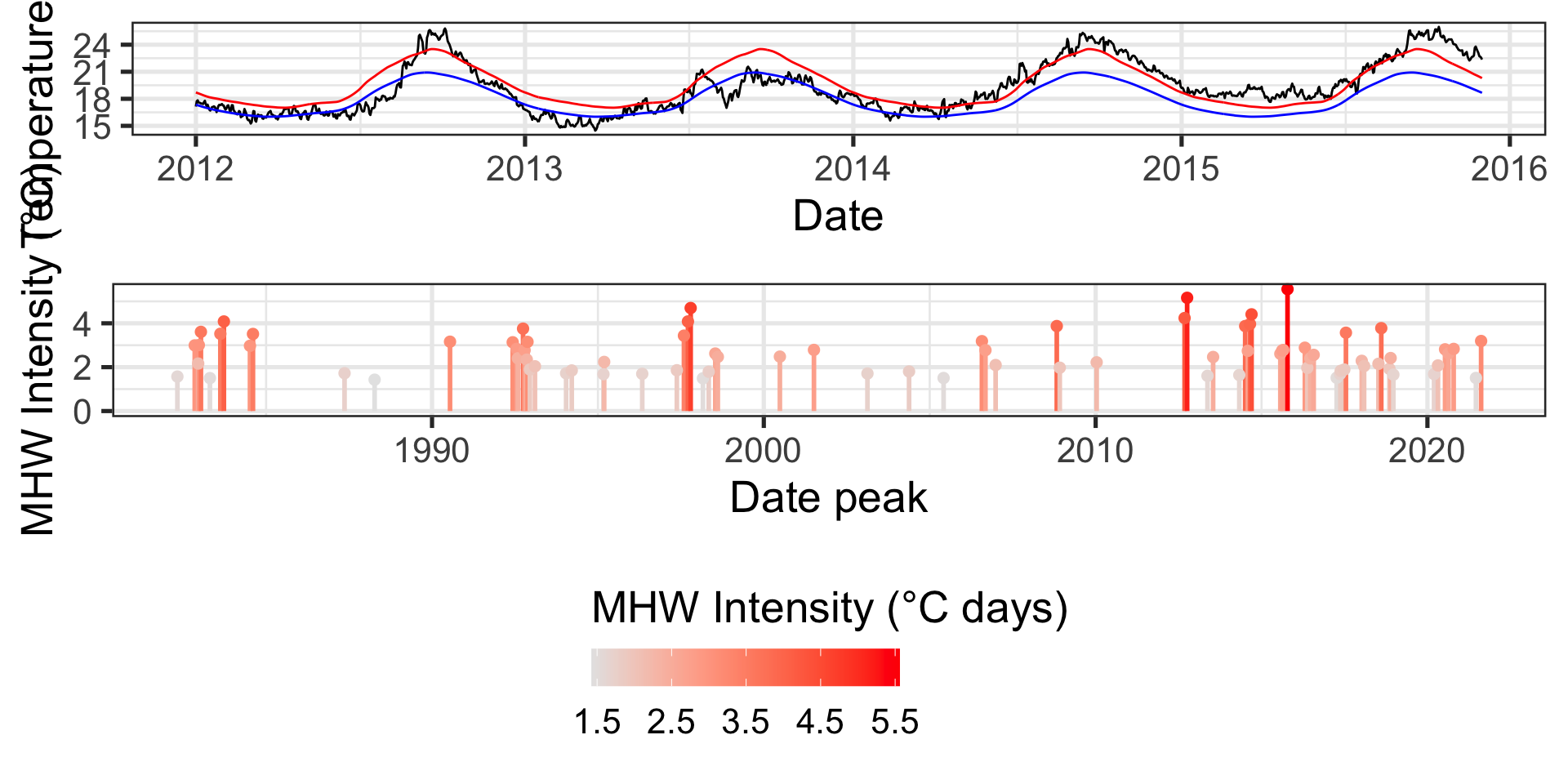
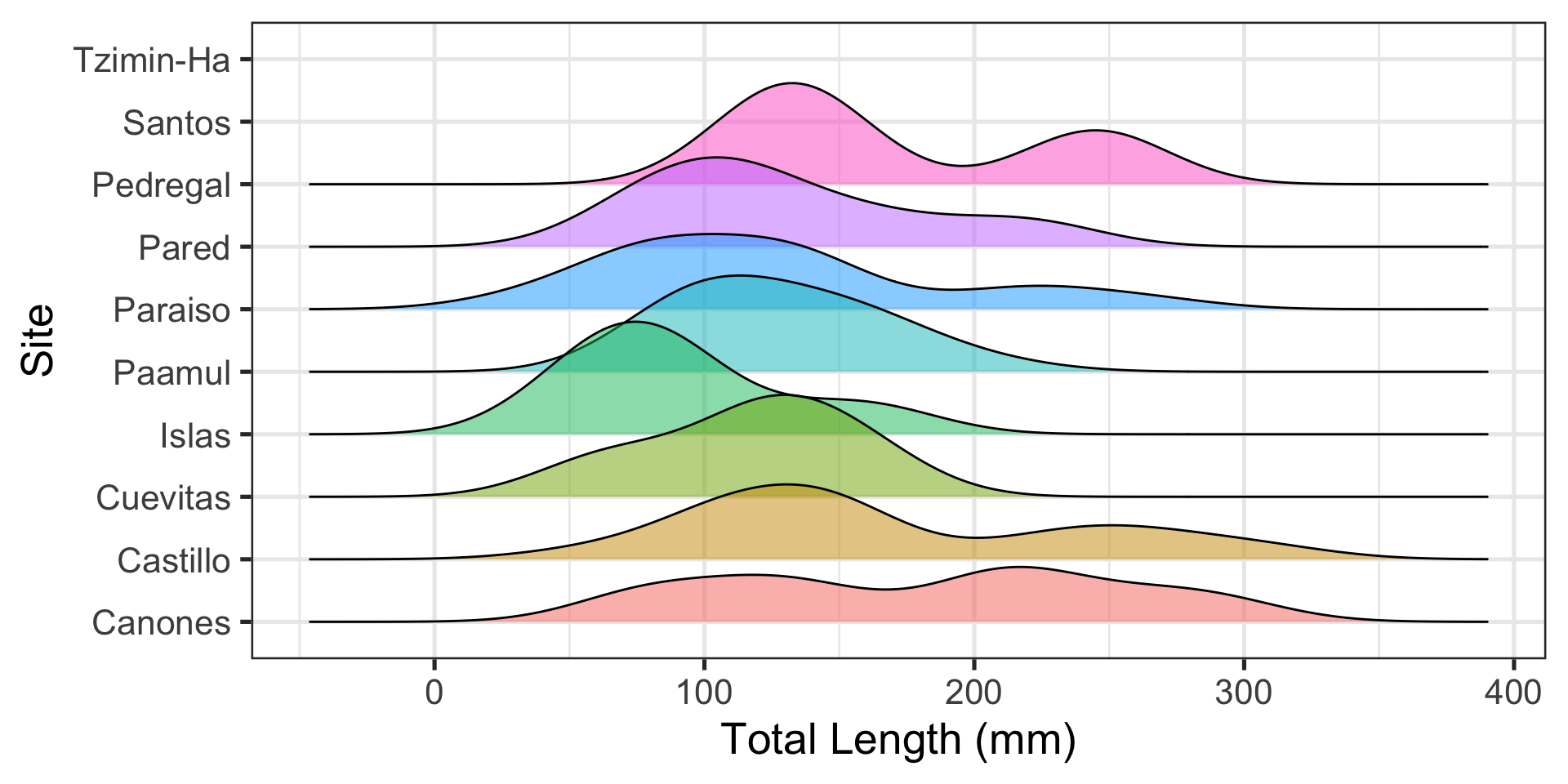
ggplot extension 2: ggridges
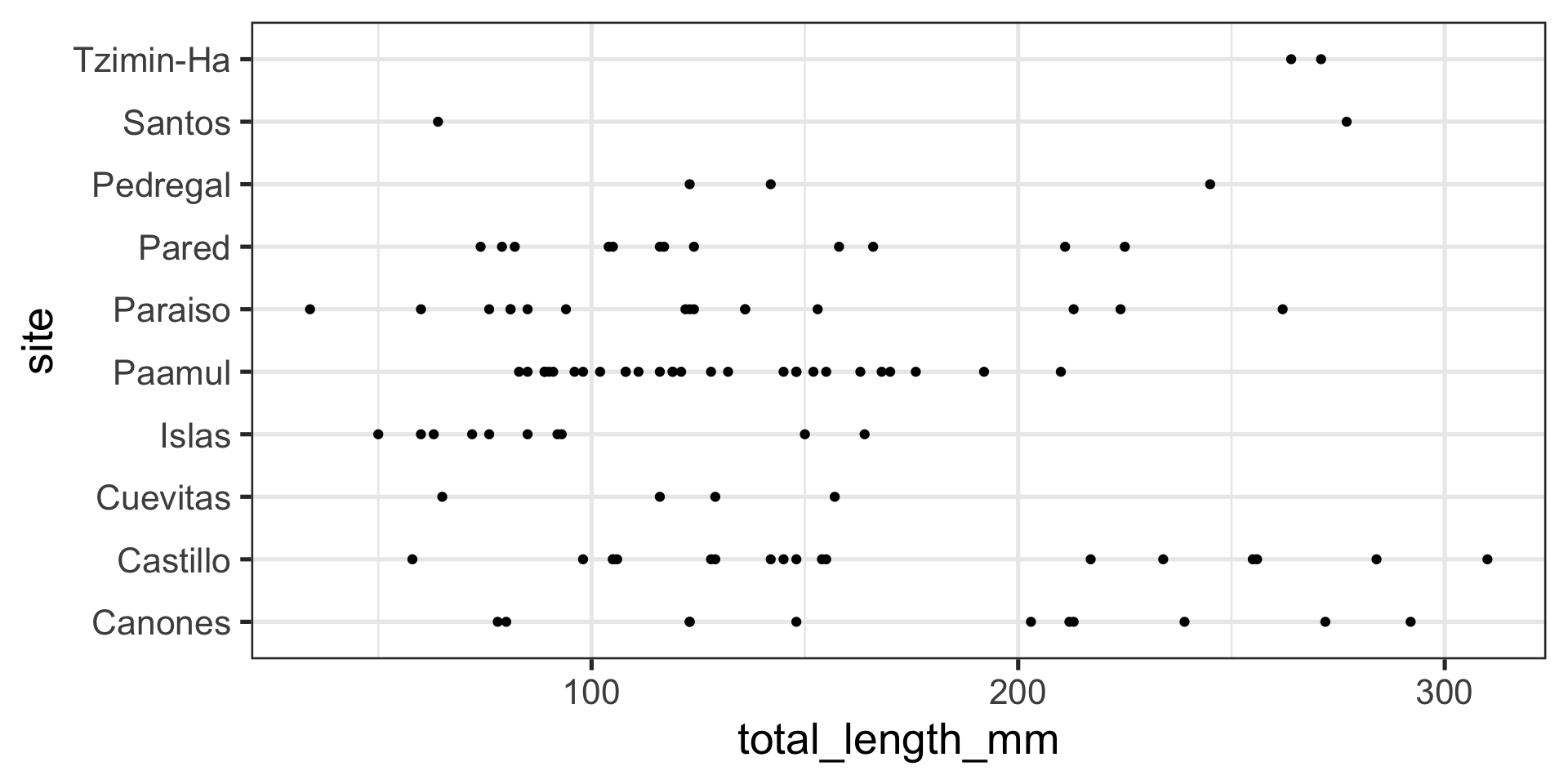
Visualizing distributions across groups is difficult
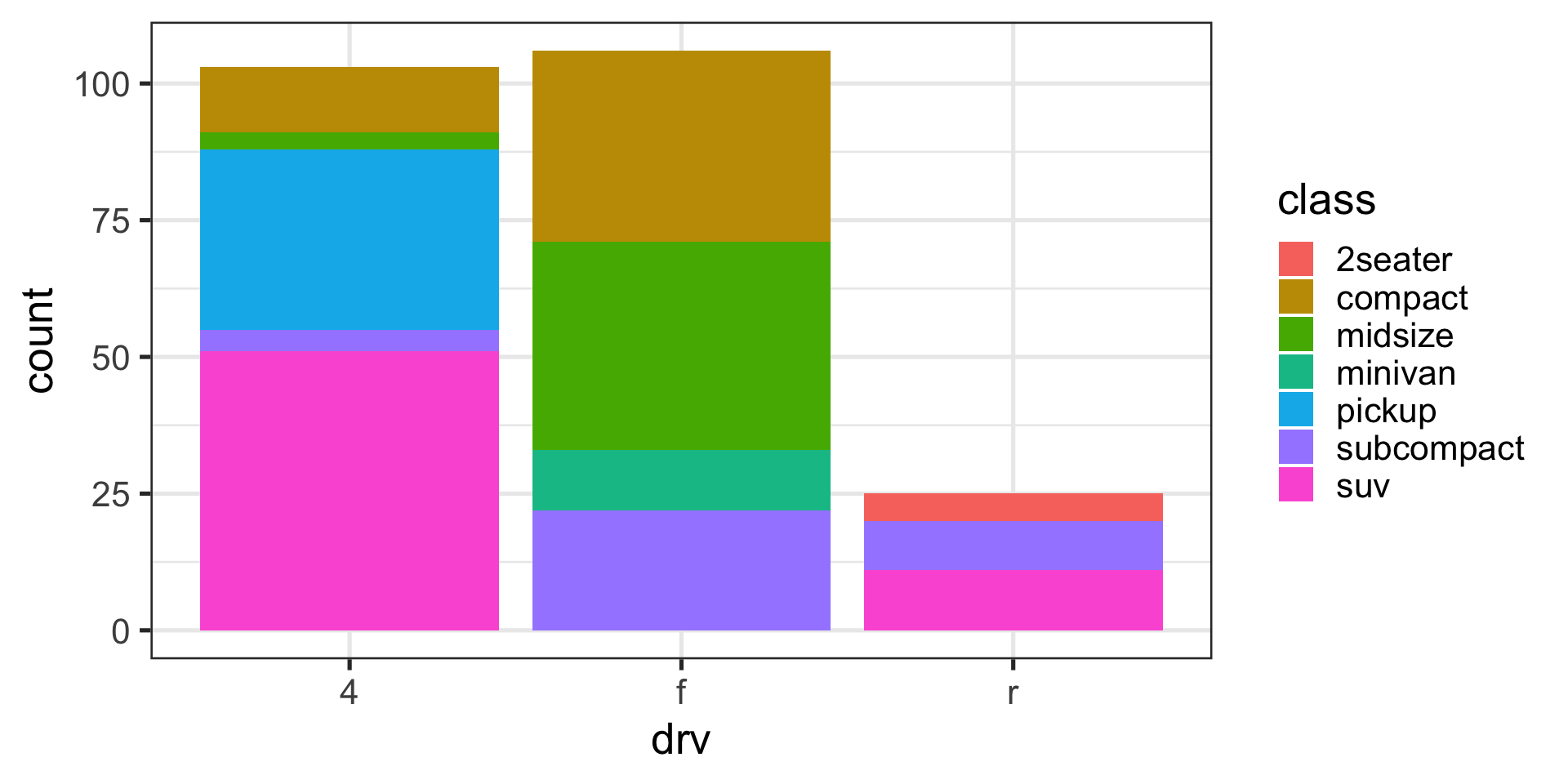
Position adjustments
Position adjustments
- Each
geomhas a defaultpositionargument - This argument controls how elements are placed on the graph
- There are four options:
- identity (allows for overlaps)
- dodge (no overlaps, move elements horizontally)
- stack (no overlaps, move elements vertically)
- fill (no overlaps, move elements vertically and fill all the space)
Position adjustments with points
Position adjustments with points
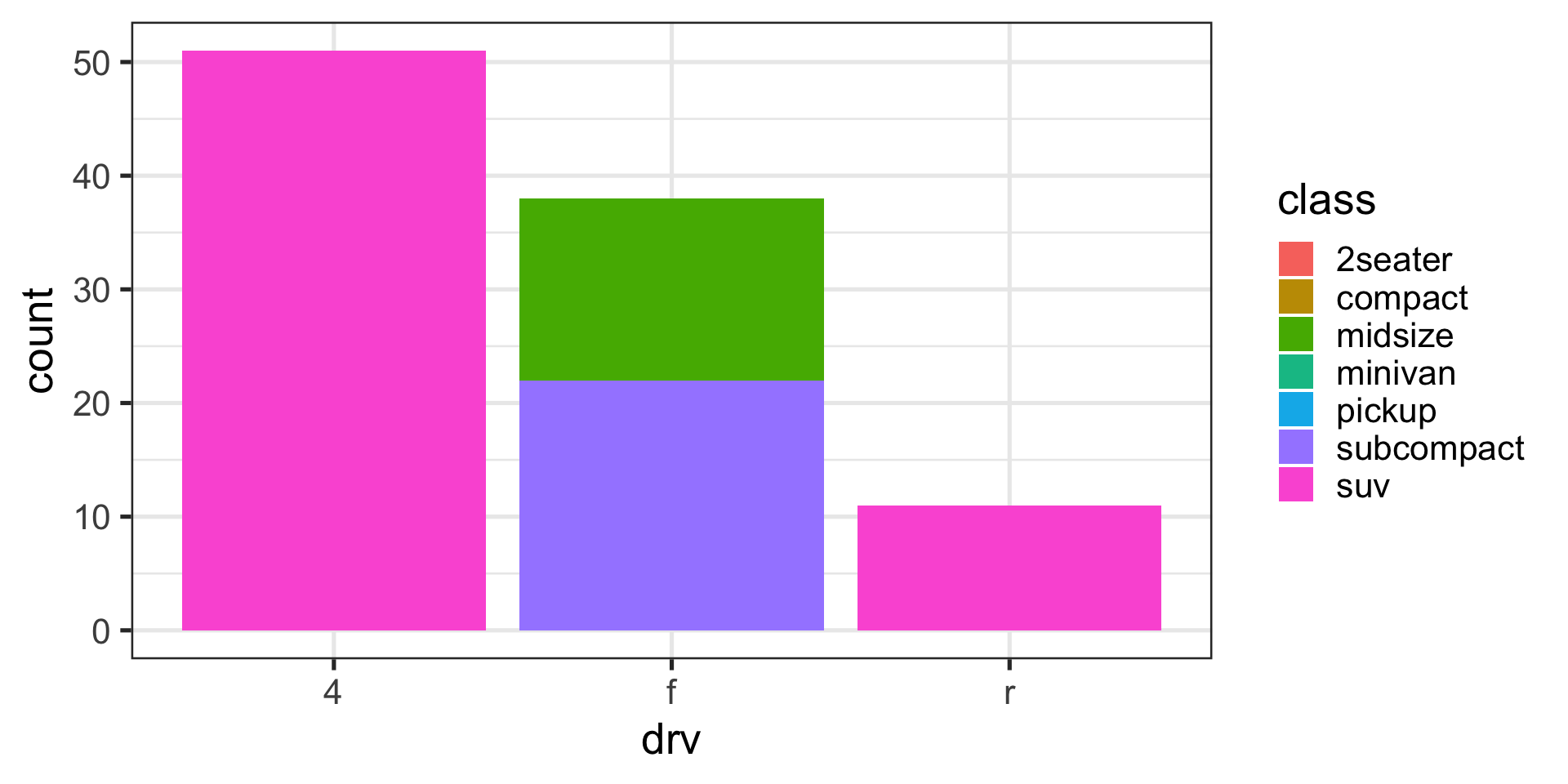
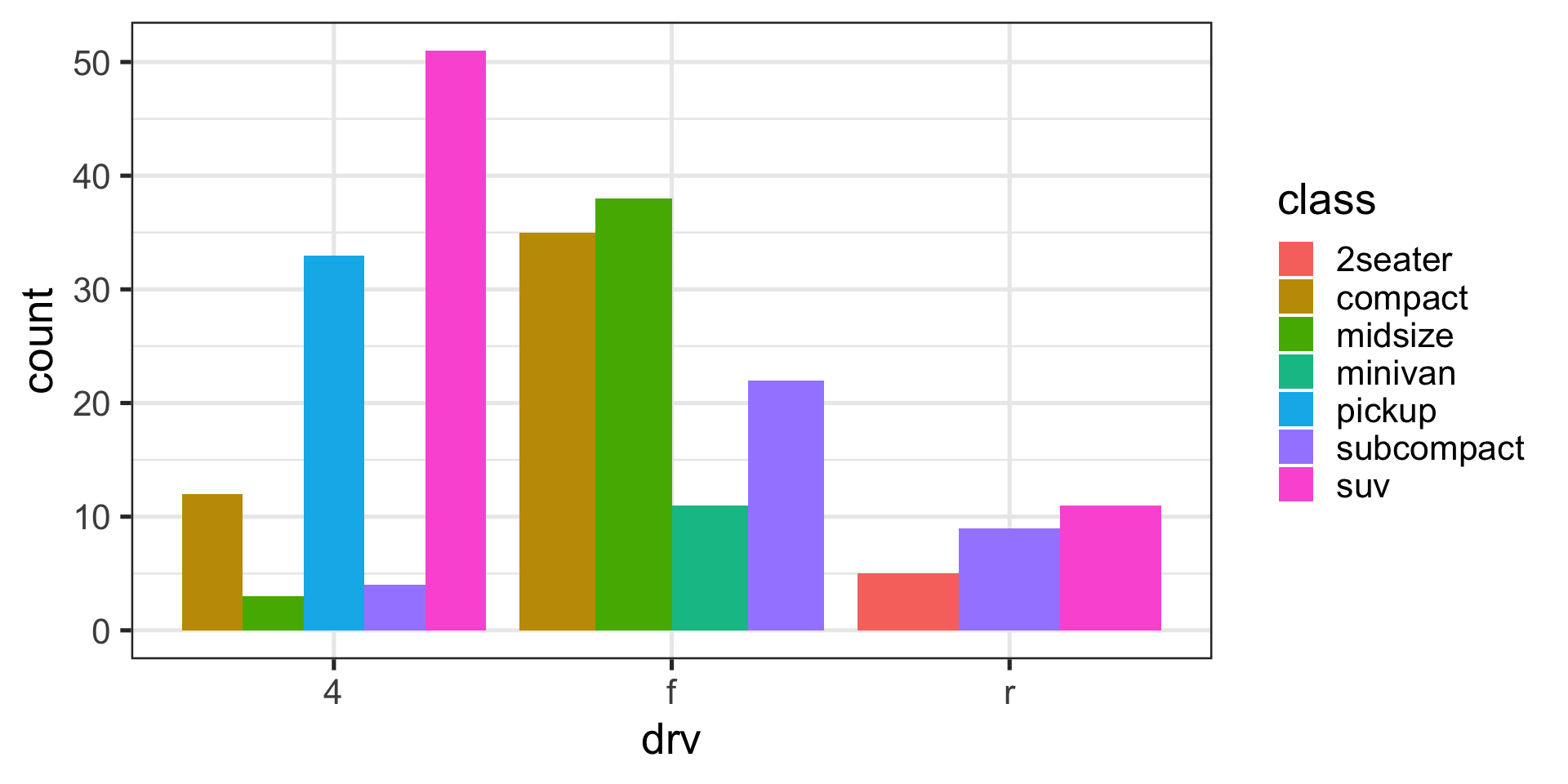
Position adjustments with bars
Stack (default)
Position adjustments with bars
Identity
Position adjustments with bars
Dodge
Position adjustments with bars
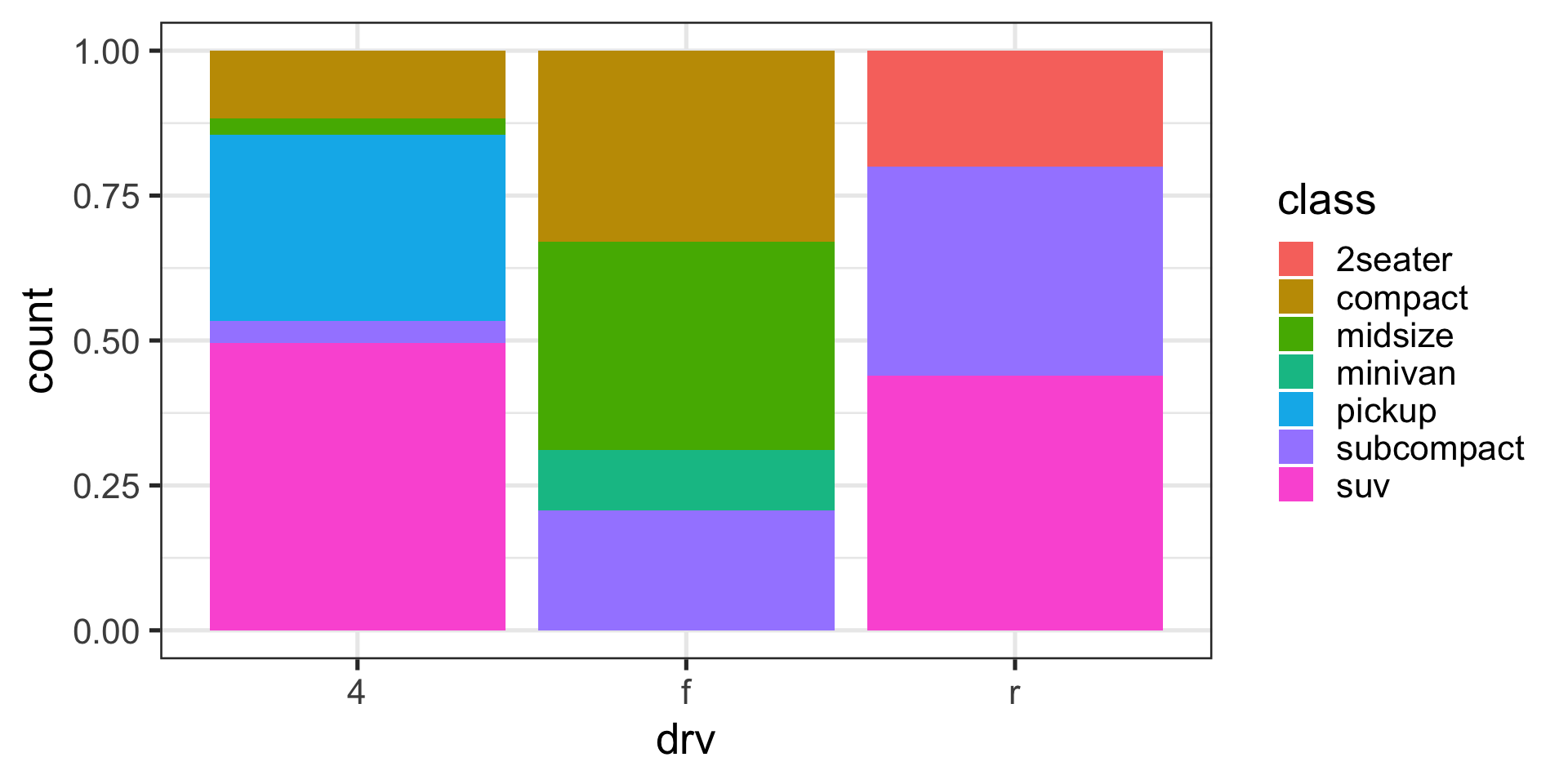
Fill
Labels
Special characters in your plots
- subindices go inside “
[]” - superscripts go after “
^” - greek letters are directly typed
- Use “
~” for spaces - See
?plotmathfor a full list
Code
data <- read_csv("https://gml.noaa.gov/webdata/ccgg/trends/co2/co2_daily_mlo.csv",
skip = 32,
col_names = c("year", "month", "day", "decimal", "co2_ppm"))
ggplot(data,
aes(x = decimal, y = co2_ppm)) +
geom_line() +
theme_minimal(base_size = 10) +
labs(x = "Date",
y = quote(CO[2]~concentration~(ppm)),
caption = "Data from the Global Monitoring Laboratory")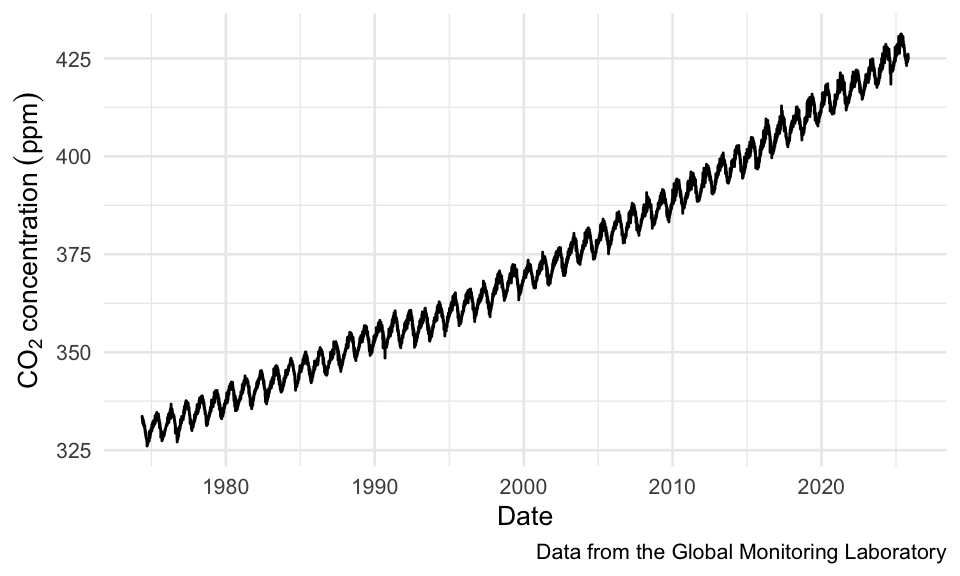
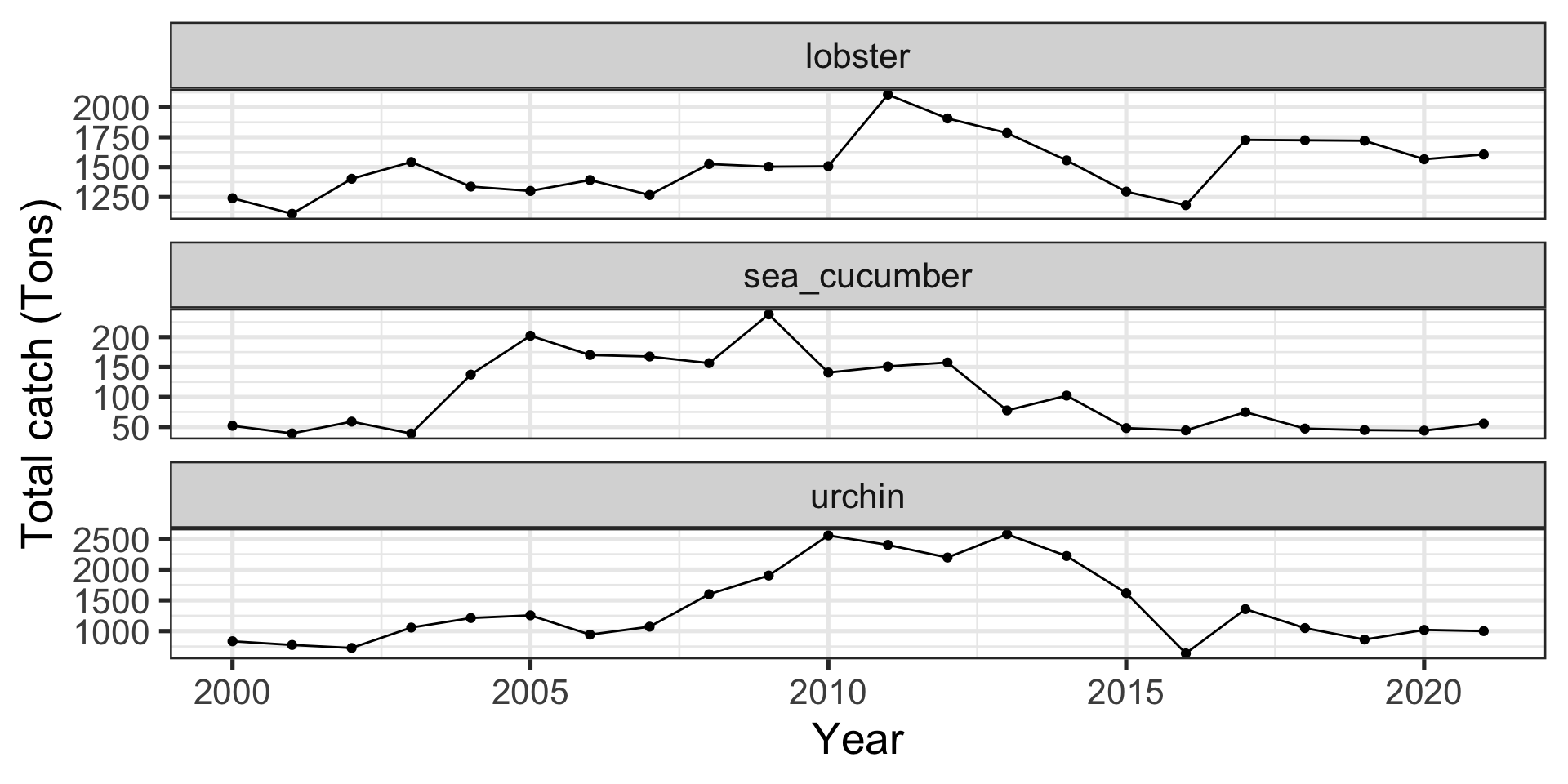
Summarizing data on the fly
Summarizing data on the fly
Sometimes you might not want to group_by and summarize, but you can go straight into a figure
Code
ggplot(data_heatwaves,
aes(x = year,
y = temp_mean)) +
stat_summary(geom = "pointrange", fun.data = "mean_se") +
stat_summary(geom = "line", fun = "mean") +
scale_x_continuous(breaks = seq(1985, 2020, by = 10)) +
facet_wrap(~str_to_sentence(str_replace(fishery, "_", " ")),
ncol = 2,
scales = "free_y") +
labs(x = "Year",
y = "Mean Temperature (°C)")
Quarto markdown
Quarto markdown
- Allows you to build documents
- slides
- html files
- pdfs
- word documents
- books…
- Particularly useful if your document heavily depends on R-generated content

Quarto markdown
- Some people use them throughout their analysis
- Pros:
- You can write tons and tons of explanations as to why you are doing something
- You can include equations
- Cons:
- You have to run the entire document, from top to bottom, every time you want to update it
- Sometimes more is not better
- Pros:
Quarto markdown
- Install Quarto markdown here
- We’ll building a document from scratch
- Then a slide
- Finally a website