install.packages("sf") # You should have this from Tuesday
install.packages("rnaturalearth") # You should have this from Tuesday
install.packages("mapview") # You should have this from Tuesday
remotes::install_github("jcvdav/EVR628tools")Working with spatial data in R: Vector
EVR 628- Intro to Environmental Data Science
Exercise 0 - Set up
- Open your
EVR628R project - Install / update the packages as needed using the code below:
- Start a new R script called
vector_data.Rand save it underscripts/02_analysis.R - Add a code header
- Load the
tidyverse,janitor,sf,rnaturalearth,mapview, andEVR628tools
Exercise 1 - Inspecting spatial data
Part 1: FL counties
Post-it up
- Go to the FDOT website and download the county Geopackage file1. Make sure you save them to your
data/rawfolder. - Look at the documentation for
?read_sf - Read the FL counties geopackage and assign it to an object called
FL_counties <- - Immediately after that, pipe into
clean_names()and then again intost_set_geometry("geometry"). FDOT data are weird, I will explain why we are doing this.
Post-it down
Code
# Load packages
library(tidyverse)
library(janitor)
library(sf)
library(rnaturalearth)
library(mapview)
library(EVR628tools)
# Load FL counties
FL_counties <- read_sf("data/raw/Florida_County_Boundaries_with_FDOT_Districts_-801746881308969010.gpkg") |>
clean_names() |>
st_set_geometry("geometry")Warning in CPL_read_ogr(dsn, layer, query, as.character(options), quiet, : GDAL
Message 1: Non-conformant content for record 1 in column CreationDate,
2025-10-07T20:05:32.0Z, successfully parsedPart 2: Inspect FL counties
Post-it up
- How many columns do we have? How many rows?
- What are the column names?
- Use R’s base
plot()to visualize them. CAREFUL, make sure you specifymax.plot = 1. - Now use
mapviewto visualize your data.
Post-it down
Code
# Inspect attributes
dim(FL_counties)[1] 67 16Code
colnames(FL_counties) [1] "name" "first_fips" "objectid_1" "fdot_distr"
[5] "district_string" "depcode" "depcodenum" "d_code"
[9] "county" "fdot_county_code" "global_id" "creation_date"
[13] "creator" "edit_date" "editor" "geometry" Code
# Base plot
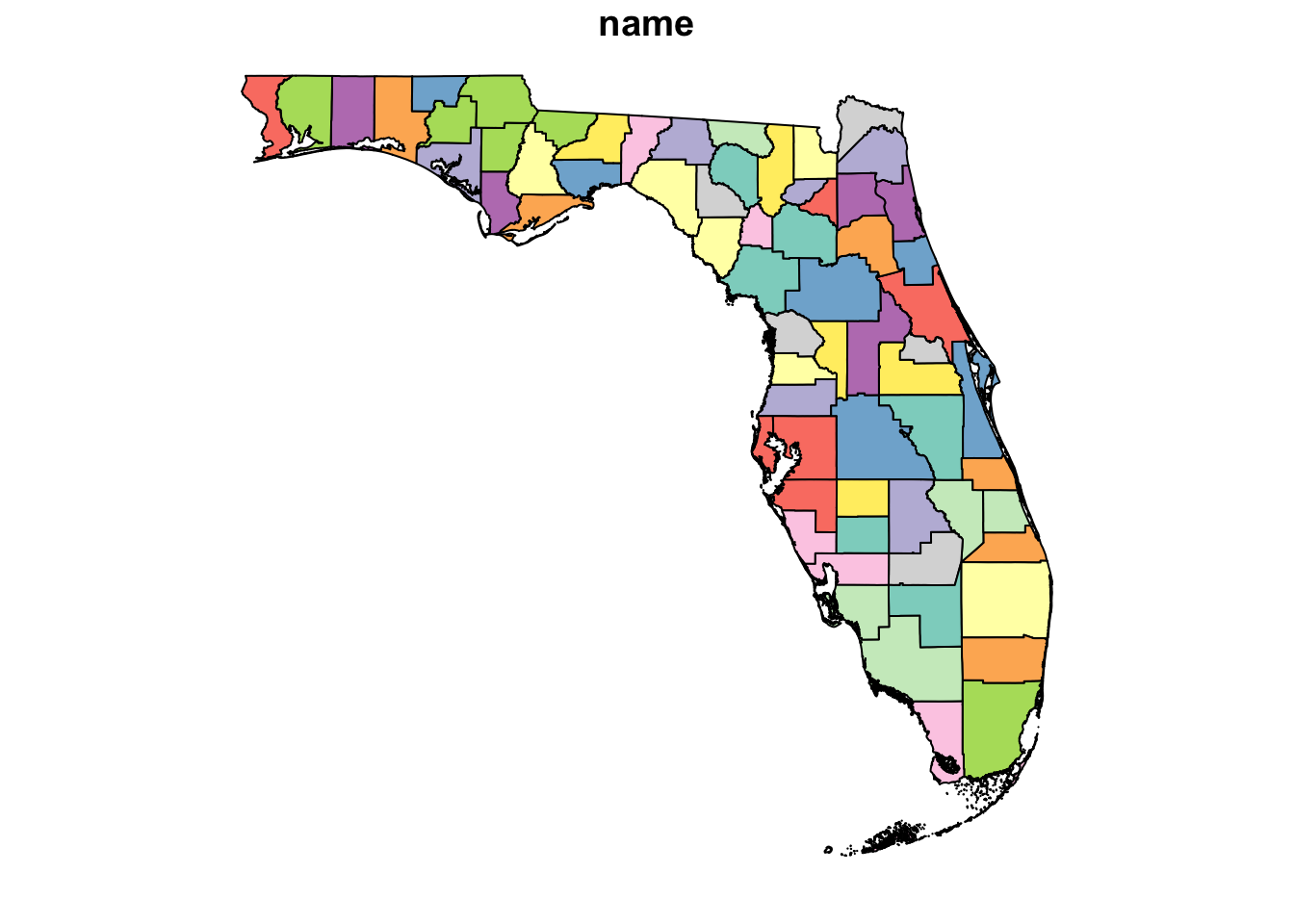
plot(FL_counties, max.plot = 1)Code
# Mapview
mapview(FL_counties)Part 3: Prep the data
Post-it up
- The FL County vector file has more columns than we need. Modify your pipeline to retain only county name and first_fips (rename them as
county_nameandcounty_fips). - Load the new
data_hurricanesdata that I’ve included in the latest version of theEVR628toolspackage - Quickly inspect them with the usual methods
Post-it down
Code
FL_counties <- read_sf("data/raw/Florida_County_Boundaries_with_FDOT_Districts_-801746881308969010.gpkg") |>
clean_names() |>
st_set_geometry("geometry") |>
select(county_name = name,
county_fips = first_fips)Warning in CPL_read_ogr(dsn, layer, query, as.character(options), quiet, : GDAL
Message 1: Non-conformant content for record 1 in column CreationDate,
2025-10-07T20:05:32.0Z, successfully parsedCode
data("data_hurricanes")
dim(data_hurricanes)[1] 51 4Code
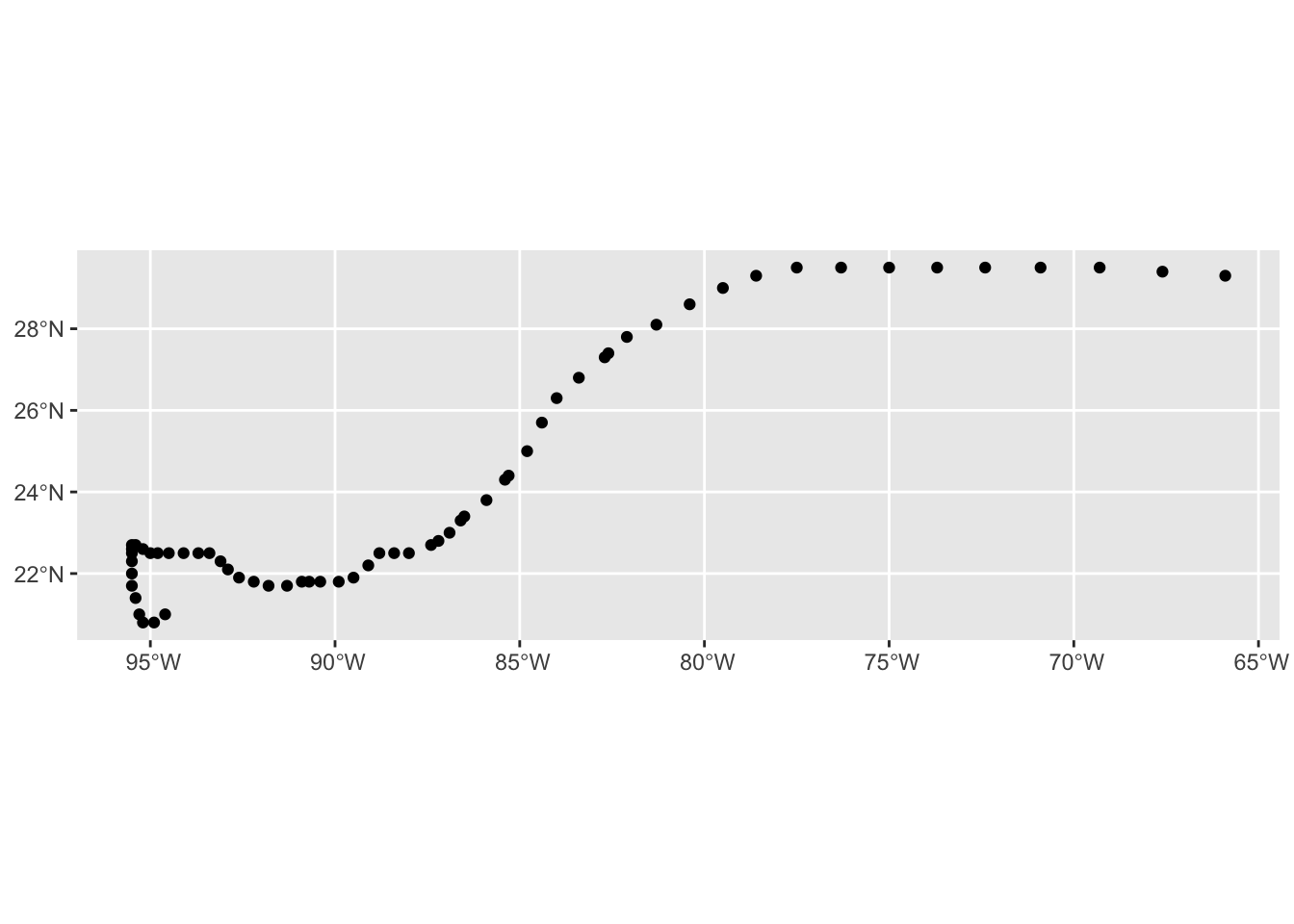
# Ignore the if statement, this is just a workaround for myself
mapview(data_hurricanes)Exercise 2 - Spatial operations
Part 1: Spatial join (st_join)
Task: Count the number of storms to which each county has been exposed between 2022 and 2024
We’ll do this one together
- Look at the documentation for the
?st_join()function - Let’s create a new object called
county_hur <-that will contain the outcome of the join operation - Now lets count the number of storms per county
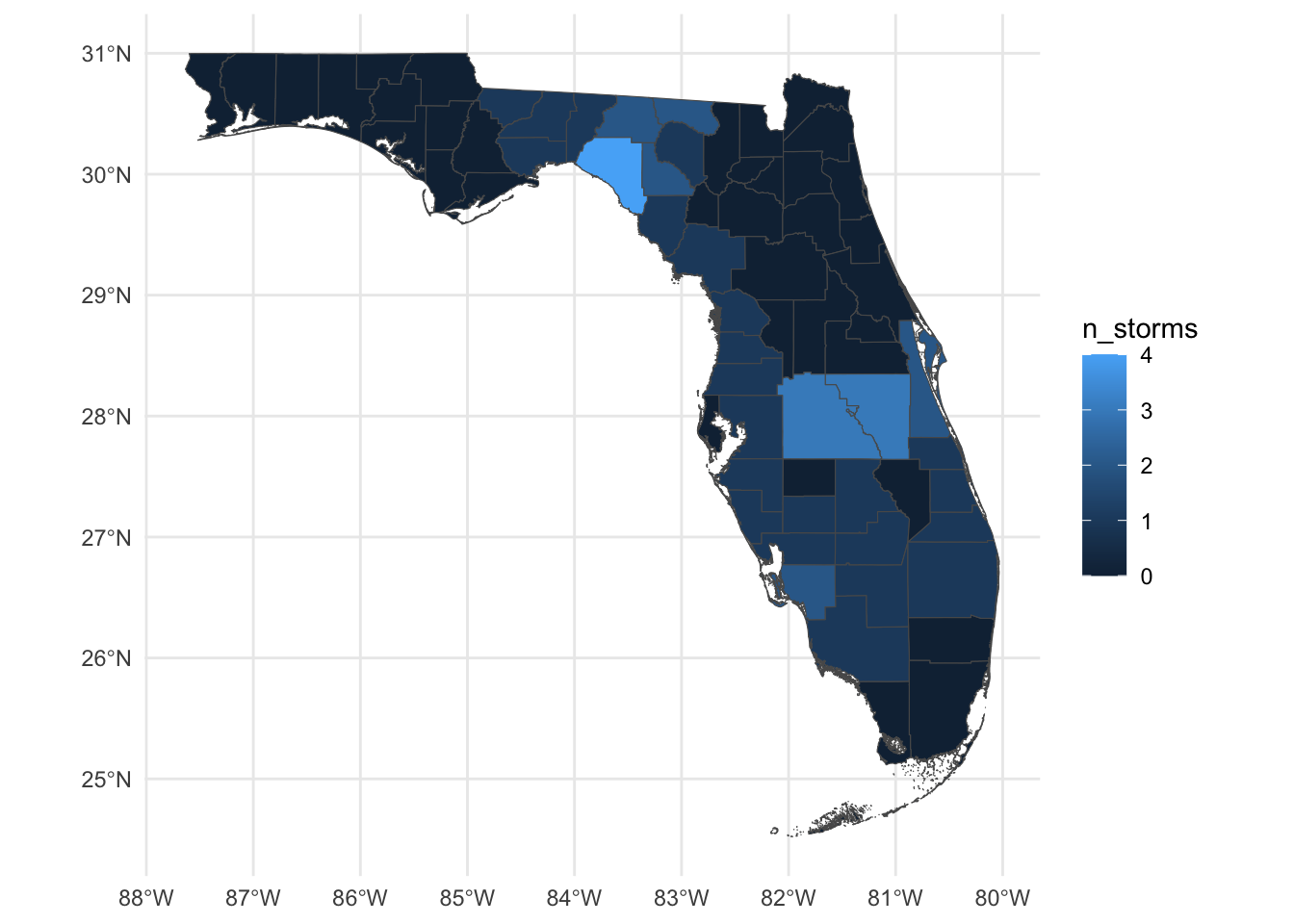
- Let’s make a map, where the fill of each county is given by the number of storms
Code
county_hur <- st_join(FL_counties, data_hurricanes) |>
group_by(county_name, county_fips) |>
summarize(n_storms = n_distinct(name,
na.rm = T))`summarise()` has grouped output by 'county_name'. You can override using the
`.groups` argument.Code
ggplot() +
geom_sf(data = county_hur,
mapping = aes(fill = n_storms)) +
theme_minimal()Part 2: Spatial filter (st_filter)
Task: Which hurricanes affected FL counties?
Post-it up
- Read the documentation for the
?st_filter()function - If our goal is to filter hurricanes based on their location, what is
xand what isy? - Use the
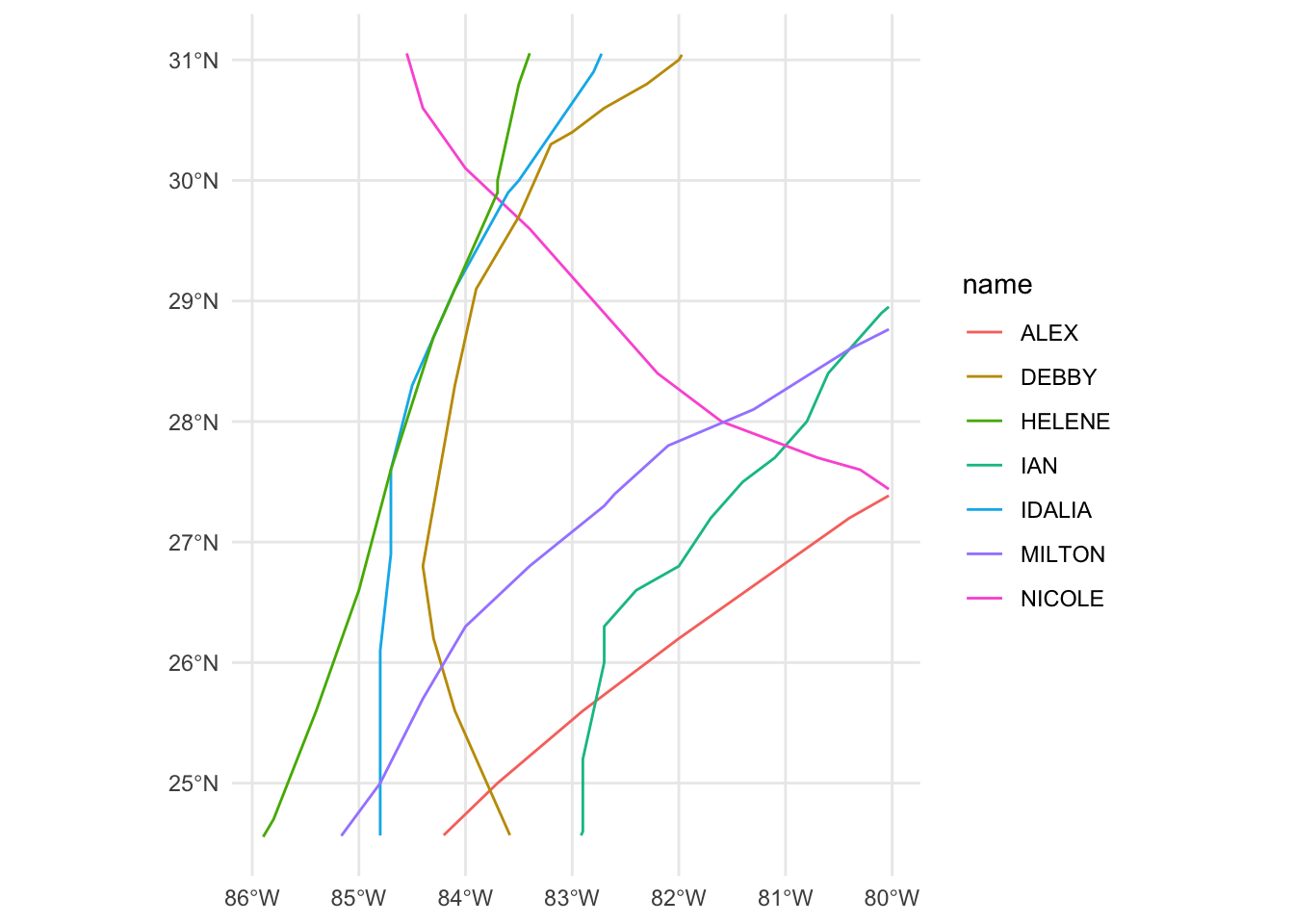
st_filter()function to find the hurricanes that went over FL counties, save the output toFL_hurricanes - Make a quick map using ggplot
- Look at the documentation for
st_crop() - Extend your pipeline for
FL_hurricanesto usest_crop()
Post-it down
Part 3: Final map
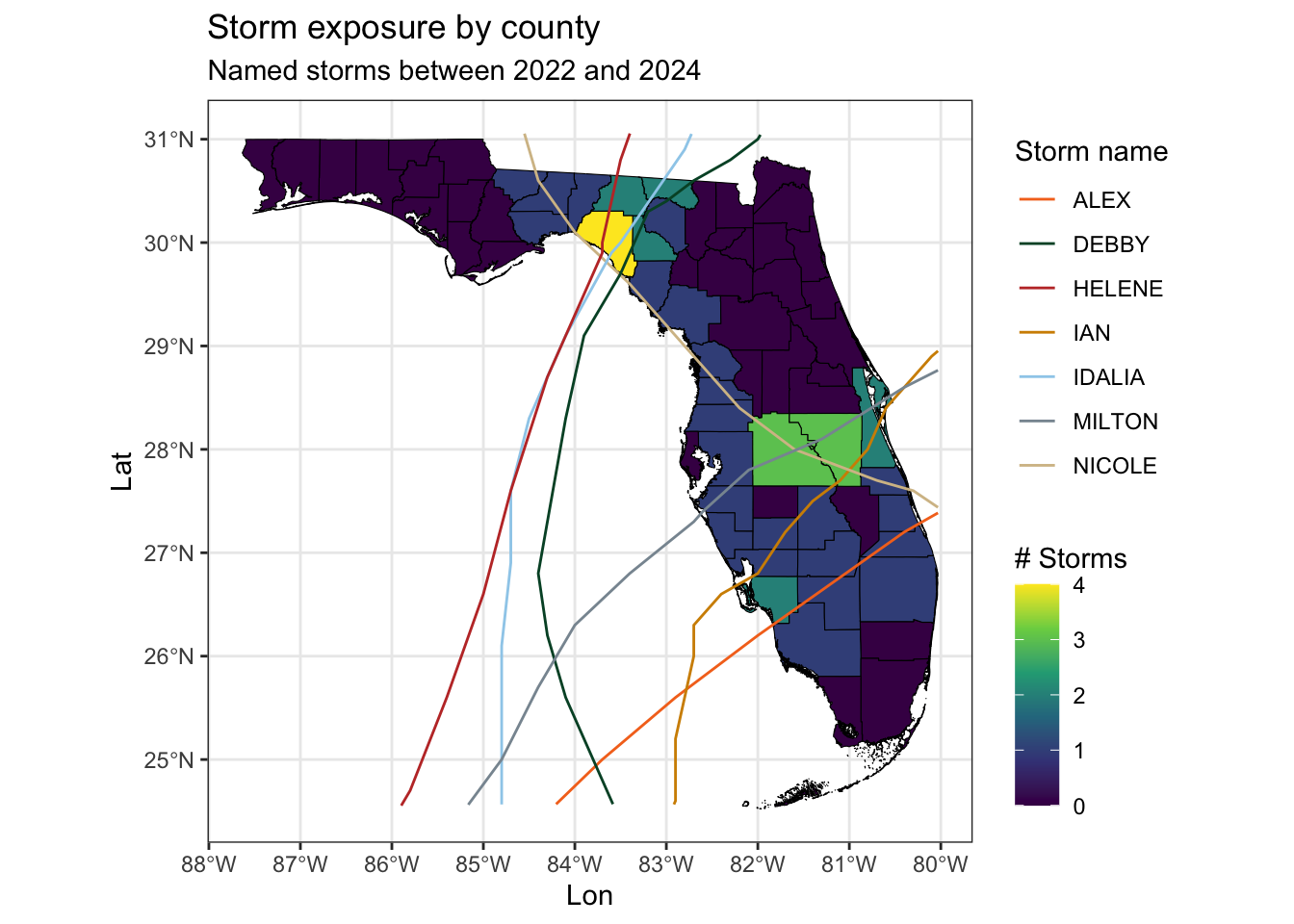
Replicate the map below
Code
ggplot() +
geom_sf(data = county_hur,
mapping = aes(fill = n_storms),
color = "black") +
geom_sf(data = FL_hurricanes,
mapping = aes(color = name)) +
scale_fill_viridis_c() +
scale_color_manual(values = palette_UM(n = 7)) +
theme_bw() +
labs(title = "Storm exposure by county",
subtitle = "Named storms between 2022 and 2024",
x = "Lon",
y = "Lat",
color = "Storm name",
fill = "# Storms")Exercise 3 - Building your own sf object
Post-it up
- Load the
data_miltondataset from theEVR628toolspackage into your environment and familiarize yourself with it (again) - Look at the documentation for
?st_as_sf() - Convert our
data_miltondata.frame into ansfobject calledmilton_sf - Plot them using base
plot()
Post-it down
Code
data("data_milton")
head(data_milton, 10)# A tibble: 10 × 7
name iso_time lat lon wind_speed pressure sshs
<chr> <dttm> <dbl> <dbl> <dbl> <dbl> <chr>
1 MILTON 2024-10-04 18:00:00 21 -94.6 30 1009 -3
2 MILTON 2024-10-04 21:00:00 20.8 -94.9 30 1009 -3
3 MILTON 2024-10-05 00:00:00 20.8 -95.2 30 1008 -3
4 MILTON 2024-10-05 03:00:00 21 -95.3 30 1008 -3
5 MILTON 2024-10-05 06:00:00 21.4 -95.4 30 1008 -3
6 MILTON 2024-10-05 09:00:00 21.7 -95.5 30 1008 -3
7 MILTON 2024-10-05 12:00:00 22 -95.5 30 1008 -1
8 MILTON 2024-10-05 15:00:00 22.3 -95.5 33 1007 -1
9 MILTON 2024-10-05 18:00:00 22.5 -95.5 35 1006 0
10 MILTON 2024-10-05 21:00:00 22.6 -95.5 35 1005 0 Code
# Convert to sf object
milton_sf <- st_as_sf(data_milton,
coords = c("lon", "lat"),
crs = "EPSG:4326")
# Plot them
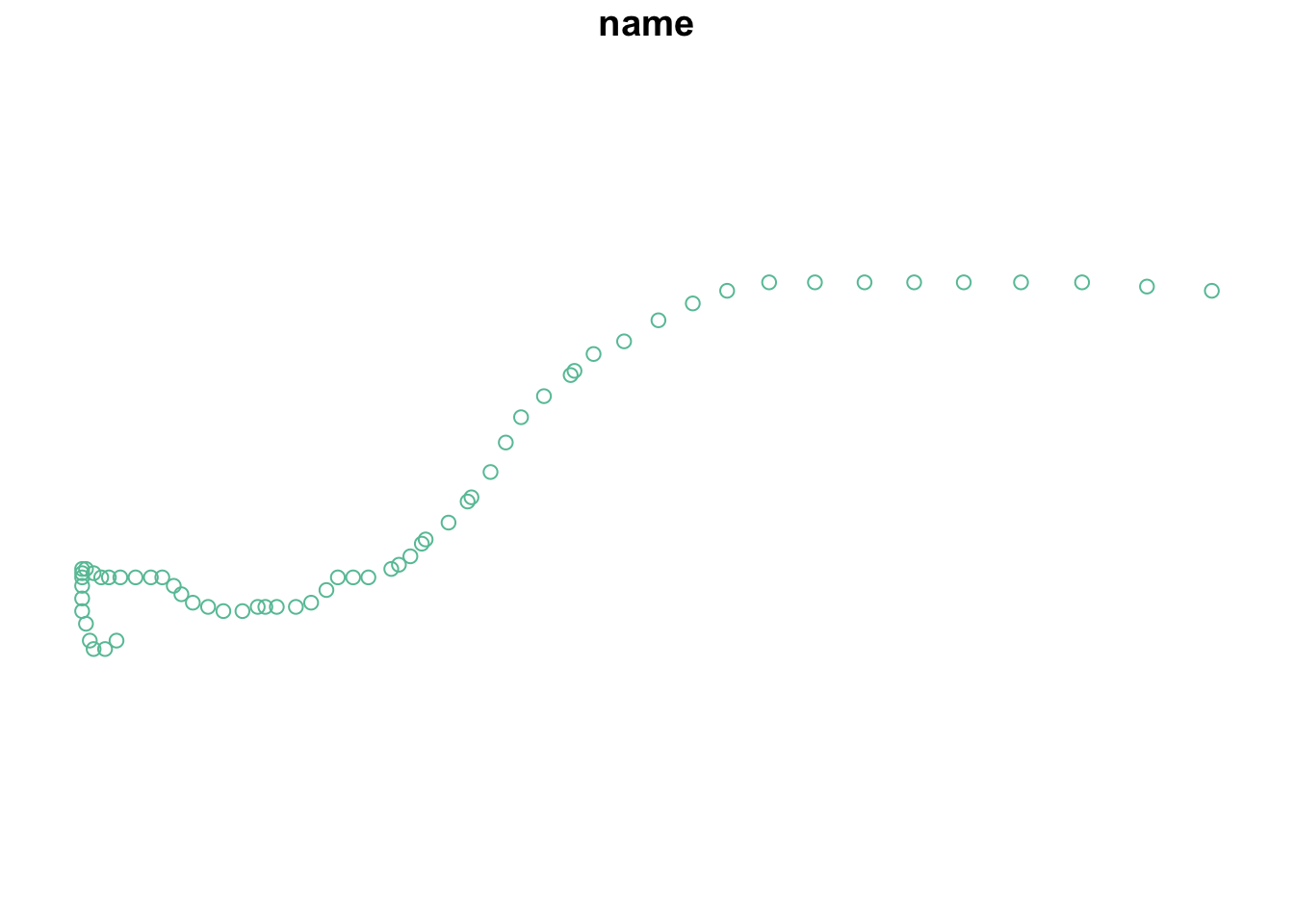
plot(milton_sf, max.plot = 1)Additional exercises
1) Converting points to linestrings
Take the milton_sf data and convert the points into a track (so, from POINT to LINESTRING)
Code
nrow(milton_sf) # One row per point[1] 62Code
milton_sf_track <- milton_sf |>
# I need to specify the groups, even though there's only one
group_by(name) |>
summarize(do_union = FALSE,
.groups = "drop")
nrow(milton_sf_track) # One row per group, but still as points in a MULTIPOINT geometry[1] 1Code
st_geometry_type(milton_sf_track)[1] MULTIPOINT
18 Levels: GEOMETRY POINT LINESTRING POLYGON MULTIPOINT ... TRIANGLECode
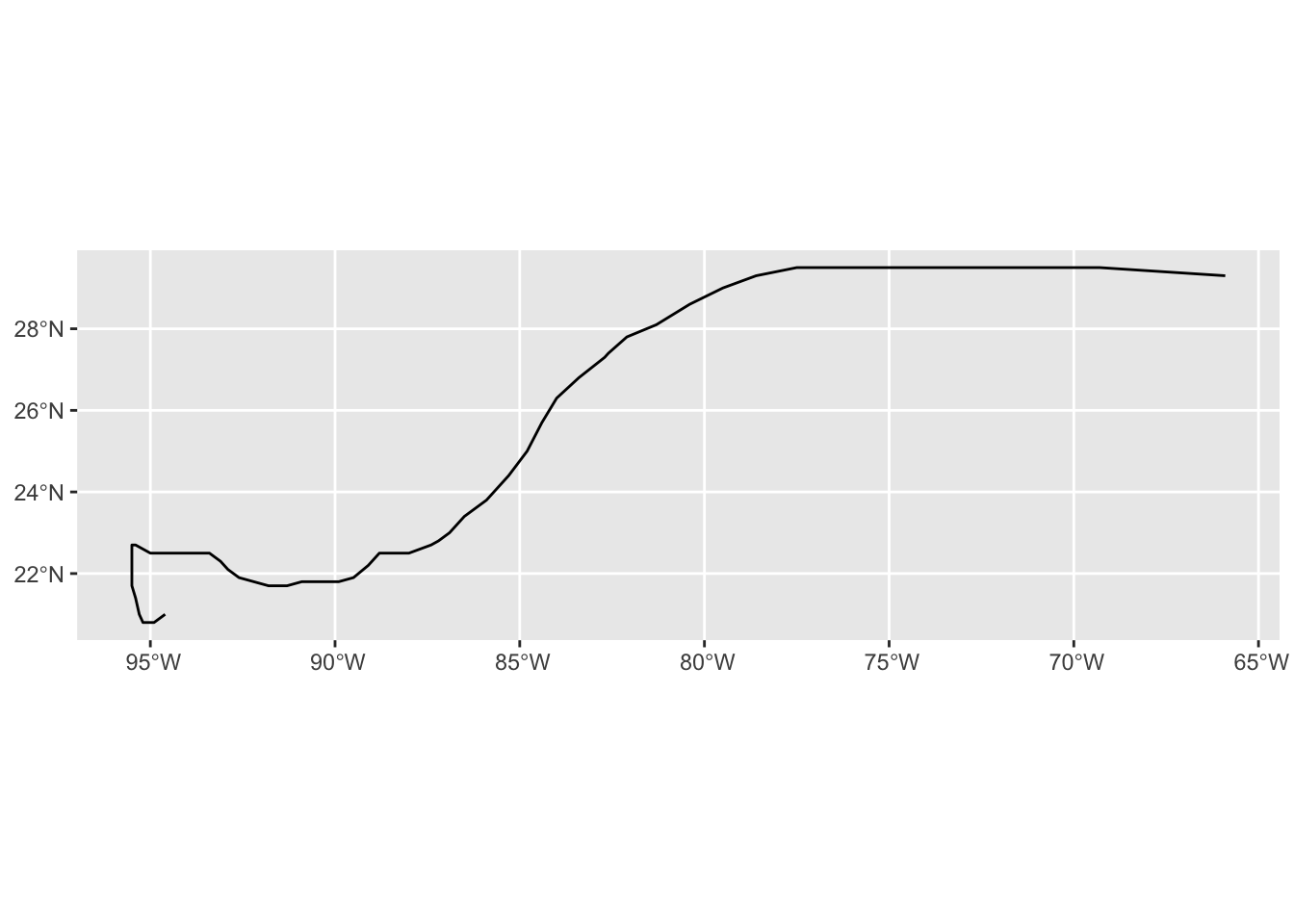
ggplot(milton_sf_track) +
geom_sf()Code
# After the summarize, all points are individually combined as a MULTIPOINT object,
# so we need to convert them to a LINESTRING. The full pipeline looks like this:
milton_sf_track <- milton_sf |>
group_by(name) |>
summarize(do_union = FALSE,
.groups = "drop") |>
st_cast("LINESTRING")
nrow(milton_sf_track) # One row per group, but now as a LINESTRING[1] 1Code
st_geometry_type(milton_sf_track)[1] LINESTRING
18 Levels: GEOMETRY POINT LINESTRING POLYGON MULTIPOINT ... TRIANGLECode
ggplot(milton_sf_track) +
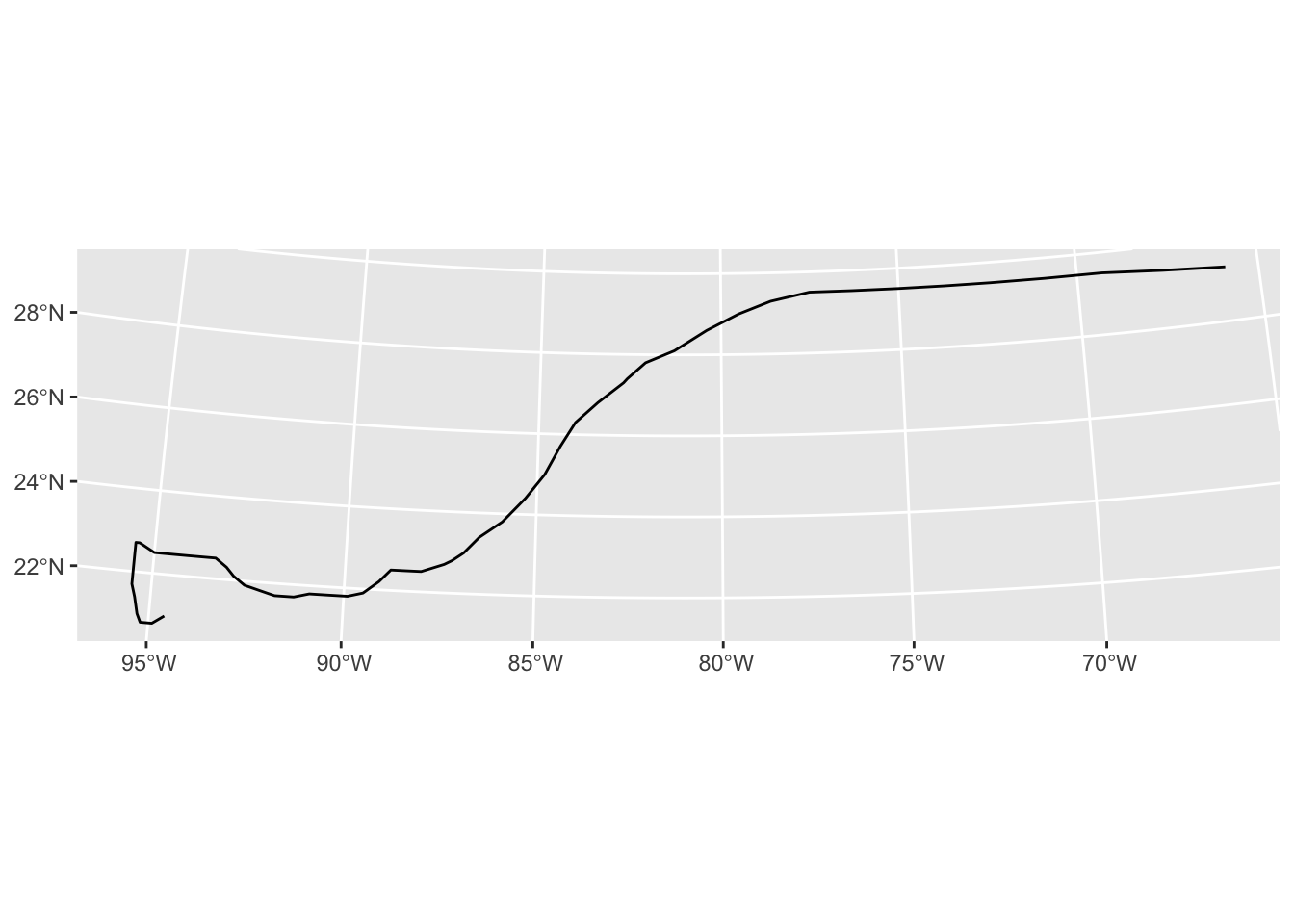
geom_sf()2) Reproject the data to a different CRS
Take the new milton_sf_track and project it to UTM zone 16N. epsg.io tells us the EPSG code is EPSG:26917-1714. After reprojecting, look at the difference in shape relative to the figure above.